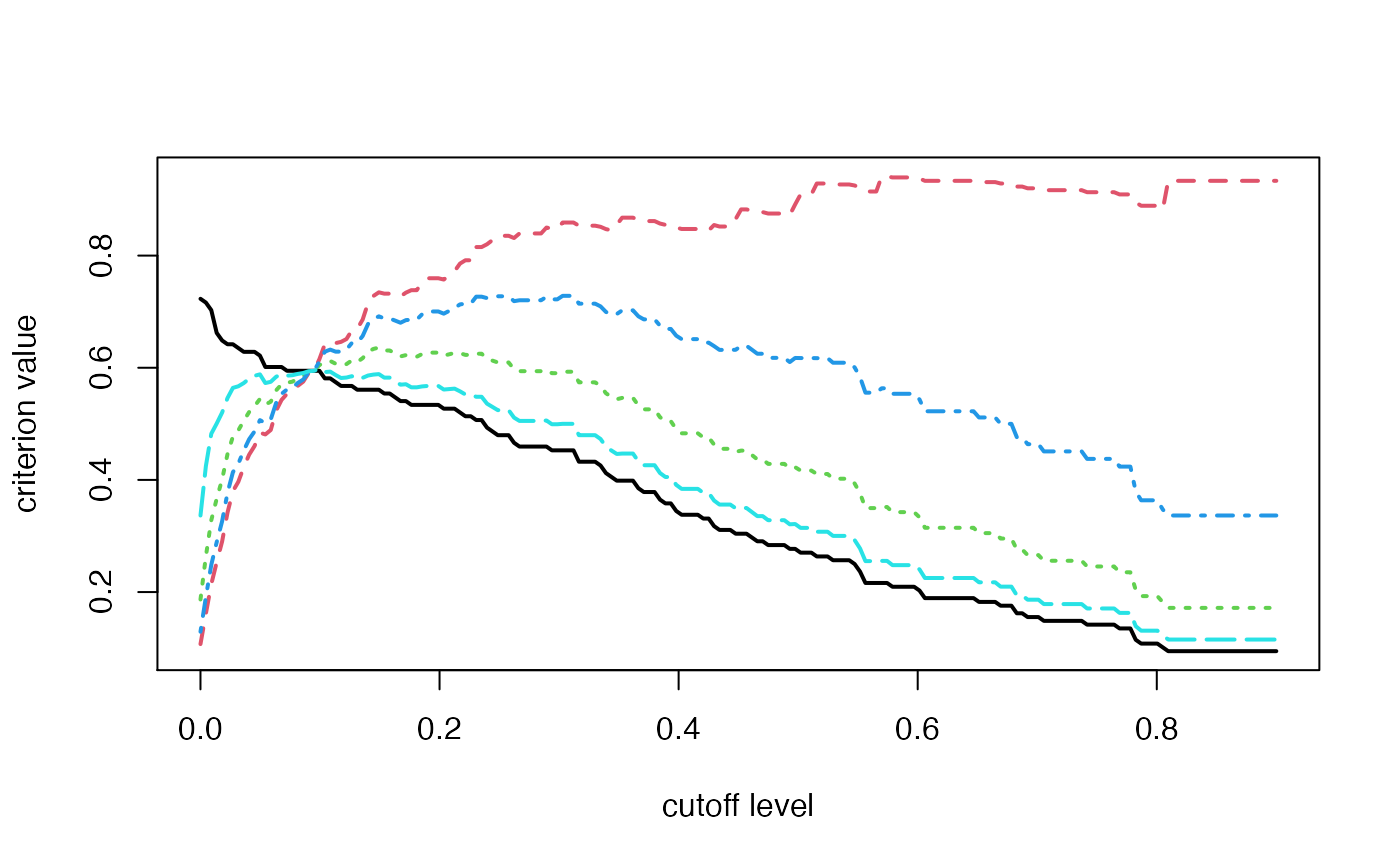
Some basic criteria of comparison between actual and inferred network.
Source:R/network.R
compare-methods.RdAllows comparison between actual and inferred network.
# S4 method for network,network,numeric compare(Net, Net_inf, nv = 1)
Arguments
| Net | A network object containing the actual network. |
|---|---|
| Net_inf | A network object containing the inferred network. |
| nv | A number that indicates at which level of cutoff the comparison should be done. |
Value
A vector containing : sensibility, predictive positive value, and the F-score
Methods
- list("signature(Net = \"network\", Net_inf = \"network\", nv = \"numeric\")")
References
Jung, N., Bertrand, F., Bahram, S., Vallat, L., and Maumy-Bertrand, M. (2014). Cascade: a R-package to study, predict and simulate the diffusion of a signal through a temporal gene network. Bioinformatics, btt705.
Vallat, L., Kemper, C. A., Jung, N., Maumy-Bertrand, M., Bertrand, F., Meyer, N., ... & Bahram, S. (2013). Reverse-engineering the genetic circuitry of a cancer cell with predicted intervention in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences, 110(2), 459-464.
Author
Nicolas Jung, Frédéric Bertrand , Myriam Maumy-Bertrand.
Examples
data(Net) data(Net_inf) #Comparing true and inferred networks F_score=NULL #Here are the cutoff level tested test.seq<-seq(0,max(abs(Net_inf@network*0.9)),length.out=200) for(u in test.seq){ F_score<-rbind(F_score,Cascade::compare(Net,Net_inf,u)) } matplot(test.seq,F_score,type="l",ylab="criterion value",xlab="cutoff level",lwd=2)