
Find the neighborhood of a set of nodes.
Source:R/omics_network.R
geneNeighborhood-omics_network-method.RdFind the neighborhood of a set of nodes.
Usage
# S4 method for class 'omics_network'
geneNeighborhood(
net,
targets,
nv = 0,
order = length(net@time_pt) - 1,
label_v = NULL,
ini = NULL,
frame.color = "white",
label.hub = FALSE,
graph = TRUE,
names = F
)Arguments
- net
a omics_network object
- targets
a vector containing the set of nodes
- nv
the level of cutoff. Defaut to 0.
- order
of the neighborhood. Defaut to `length(net@time_pt)-1`.
- label_v
vector defining the vertex labels.
- ini
using the “position” function, you can fix the position of the nodes.
- frame.color
color of the frames.
- label.hub
logical ; if TRUE only the hubs are labeled.
- graph
plot graph of the network. Defaults to `TRUE`.
- names
return names of the neighbors. Defaults to `FALSE`.
Examples
data(Selection)
data(infos)
#Find probesets for EGR1
pbst_EGR1 = infos[infos$hgnc_symbol=="EGR1", "affy_hg_u133_plus_2"]
gene_IDs = infos[match(Selection@name, infos$affy_hg_u133_plus_), "hgnc_symbol"]
data(network)
#A nv value can chosen using the cutoff function
nv=.11
EGR1<-which(is.element(Selection@name,pbst_EGR1))
P<-position(network,nv=nv)
geneNeighborhood(network,targets=EGR1,nv=nv,ini=P,
label_v=gene_IDs)
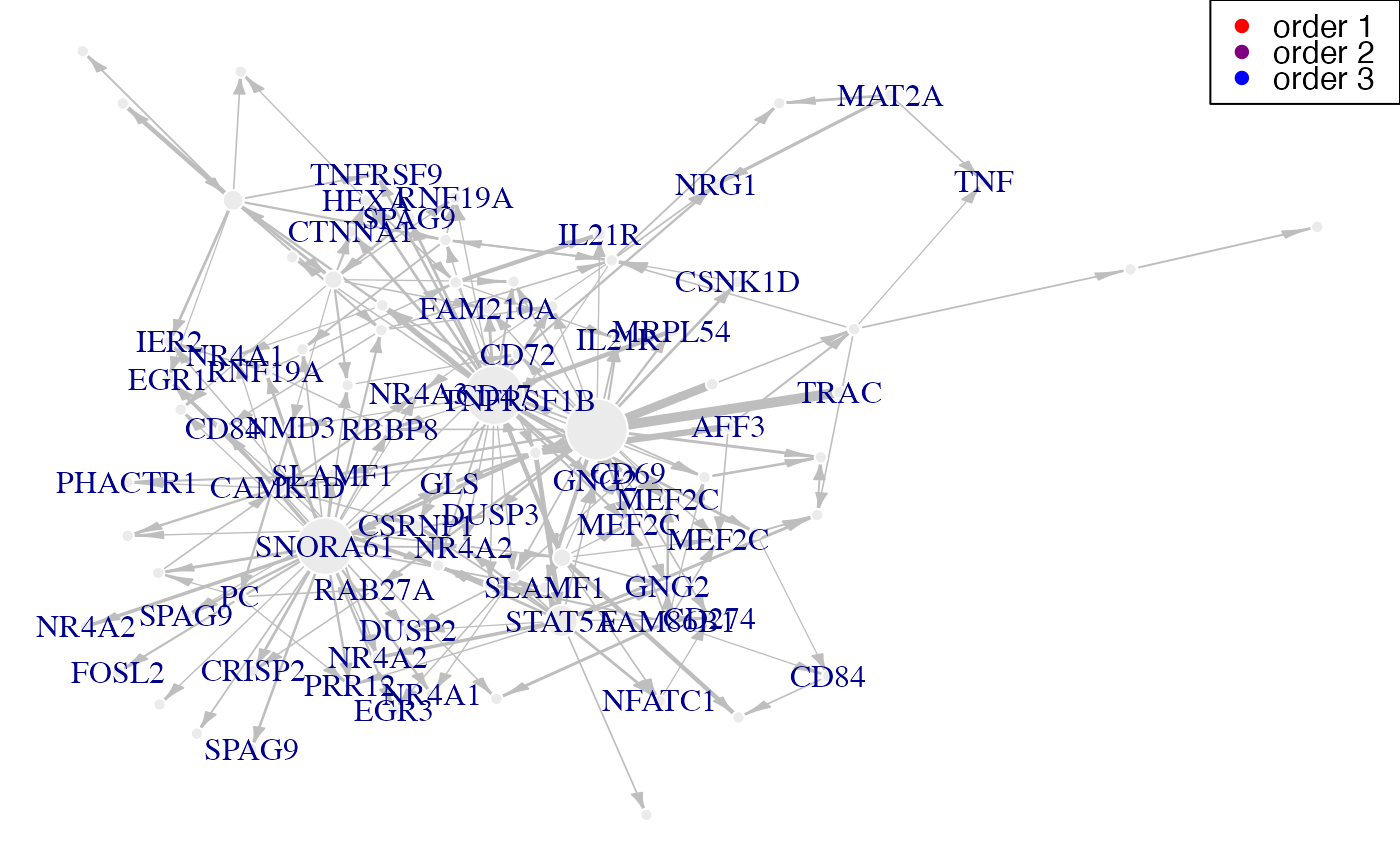 #> [[1]]
#> [[1]][[1]]
#> + 1/102 vertex, from e394b8d:
#> [1] 25
#>
#> [[1]][[2]]
#> + 1/102 vertex, from e394b8d:
#> [1] 26
#>
#>
#> [[2]]
#> [[2]][[1]]
#> + 1/102 vertex, from e394b8d:
#> [1] 25
#>
#> [[2]][[2]]
#> + 1/102 vertex, from e394b8d:
#> [1] 26
#>
#>
#> [[3]]
#> [[3]][[1]]
#> + 1/102 vertex, from e394b8d:
#> [1] 25
#>
#> [[3]][[2]]
#> + 1/102 vertex, from e394b8d:
#> [1] 26
#>
#>
#> [[1]]
#> [[1]][[1]]
#> + 1/102 vertex, from e394b8d:
#> [1] 25
#>
#> [[1]][[2]]
#> + 1/102 vertex, from e394b8d:
#> [1] 26
#>
#>
#> [[2]]
#> [[2]][[1]]
#> + 1/102 vertex, from e394b8d:
#> [1] 25
#>
#> [[2]][[2]]
#> + 1/102 vertex, from e394b8d:
#> [1] 26
#>
#>
#> [[3]]
#> [[3]][[1]]
#> + 1/102 vertex, from e394b8d:
#> [1] 25
#>
#> [[3]][[2]]
#> + 1/102 vertex, from e394b8d:
#> [1] 26
#>
#>