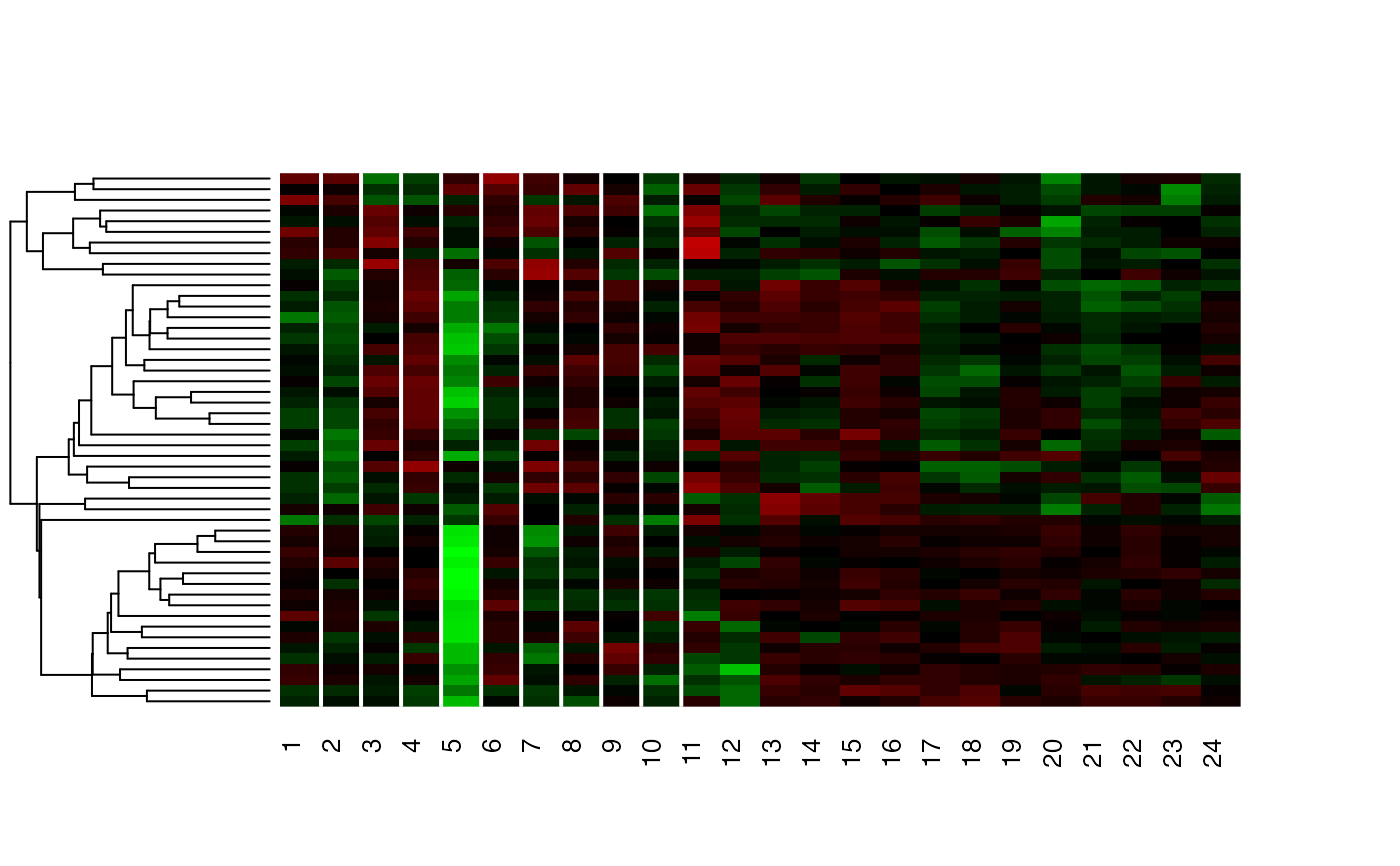
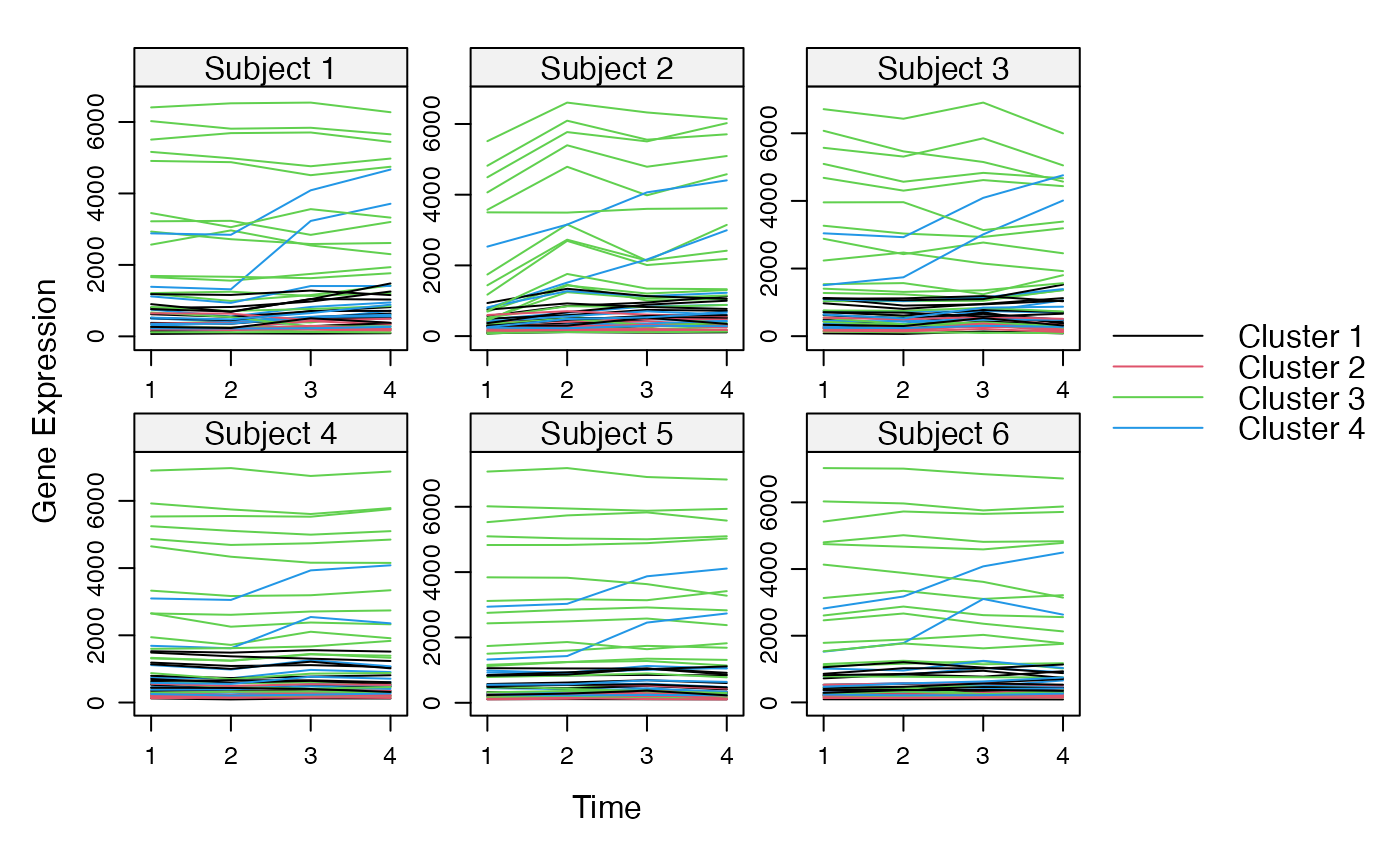
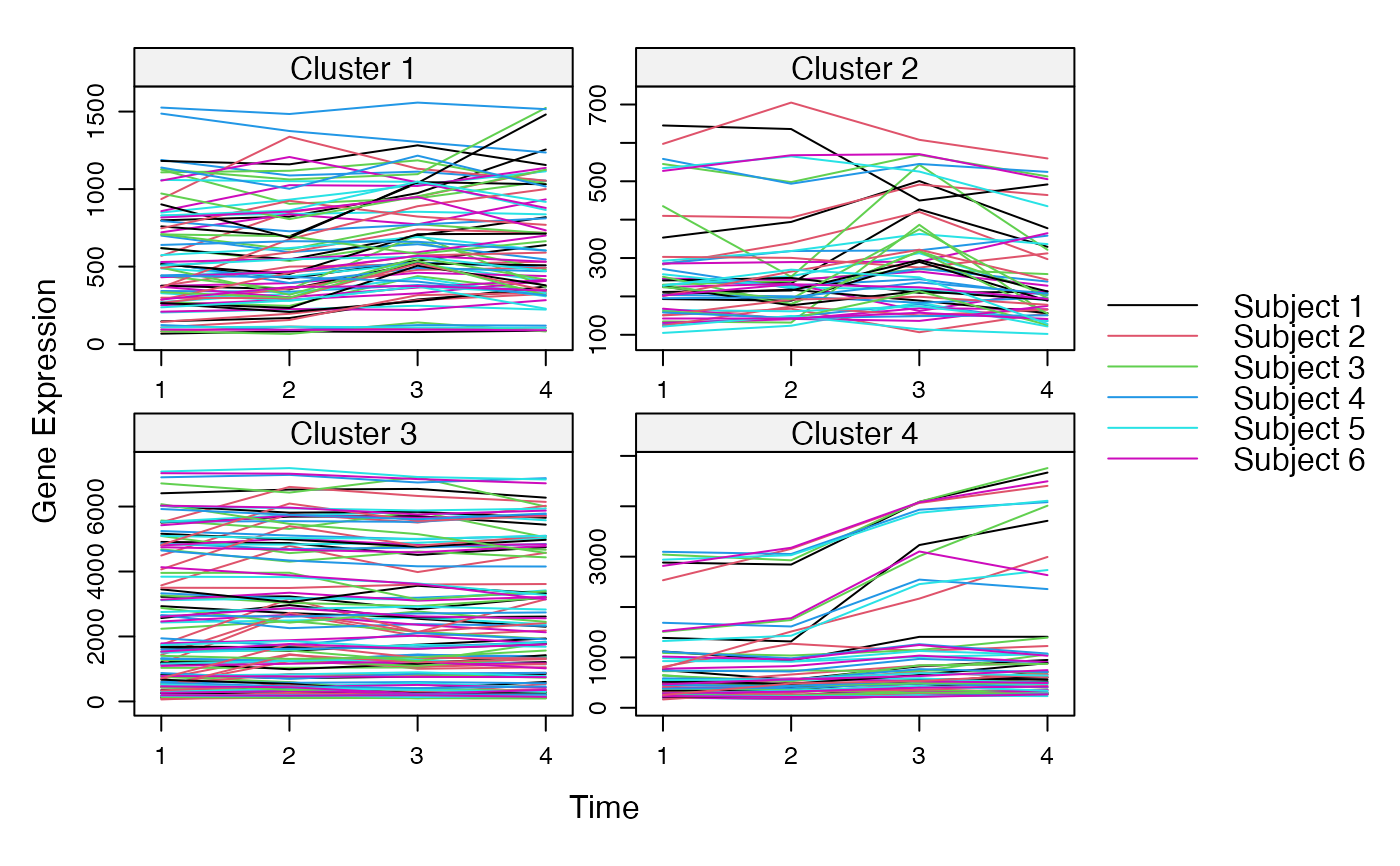
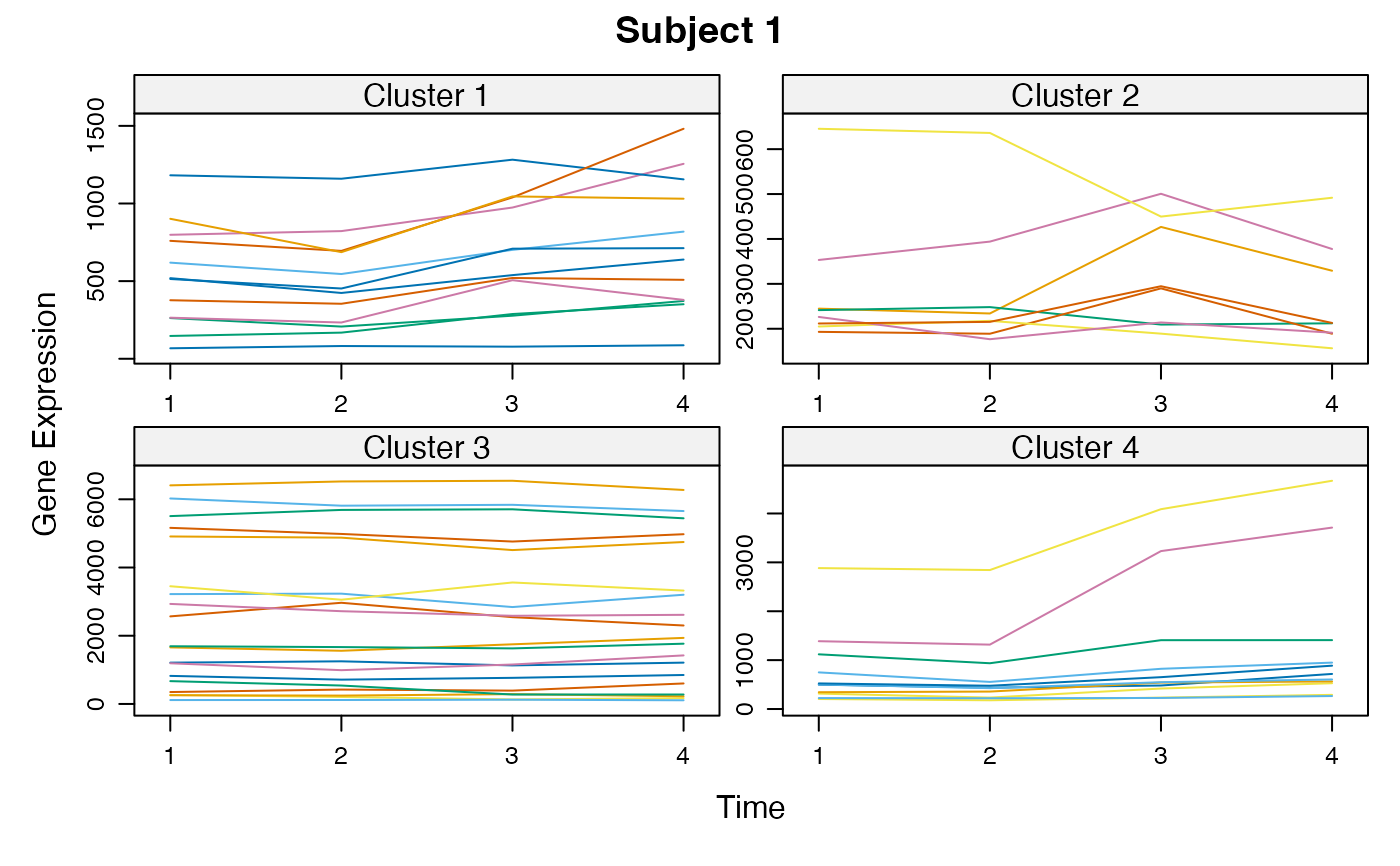
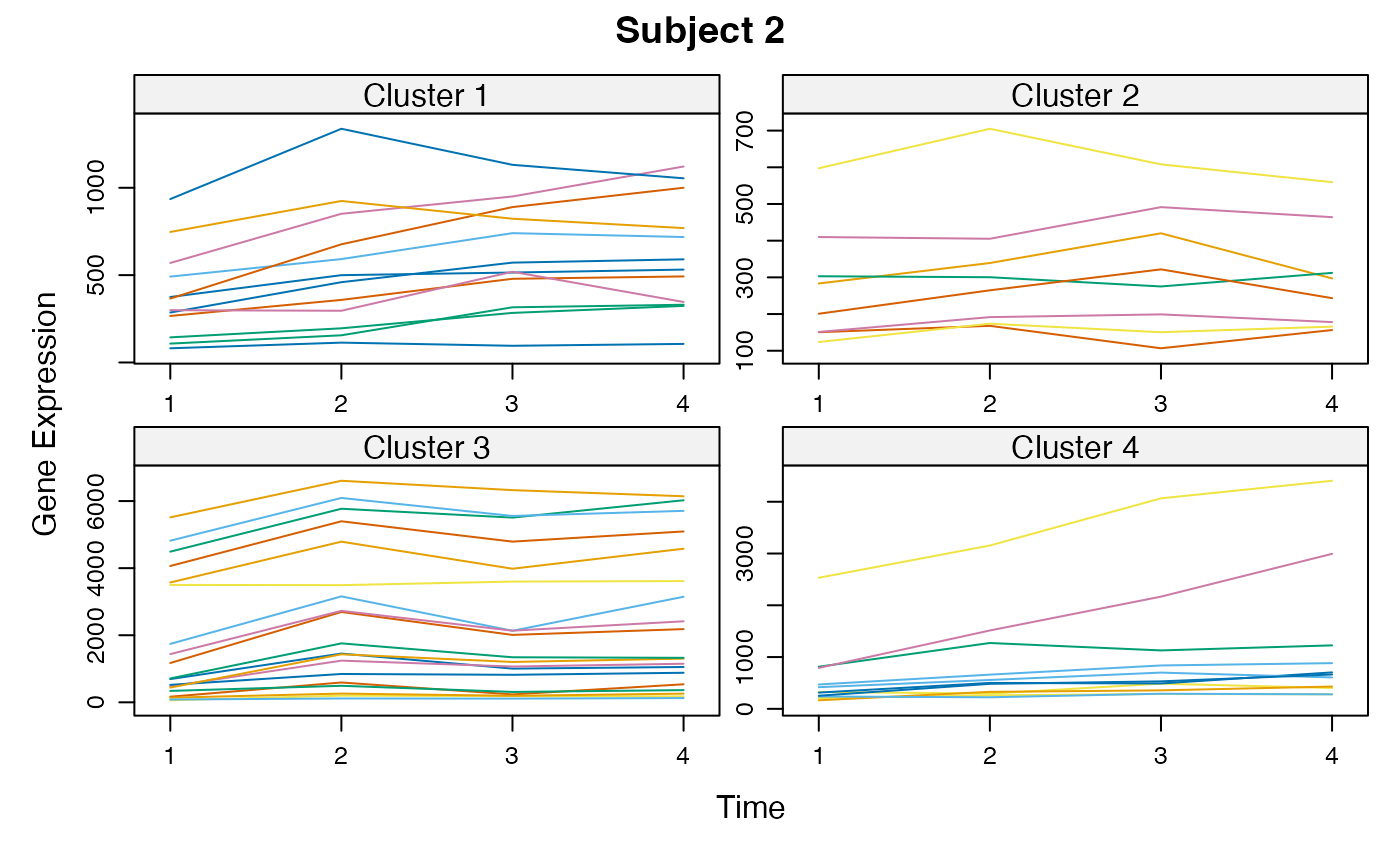
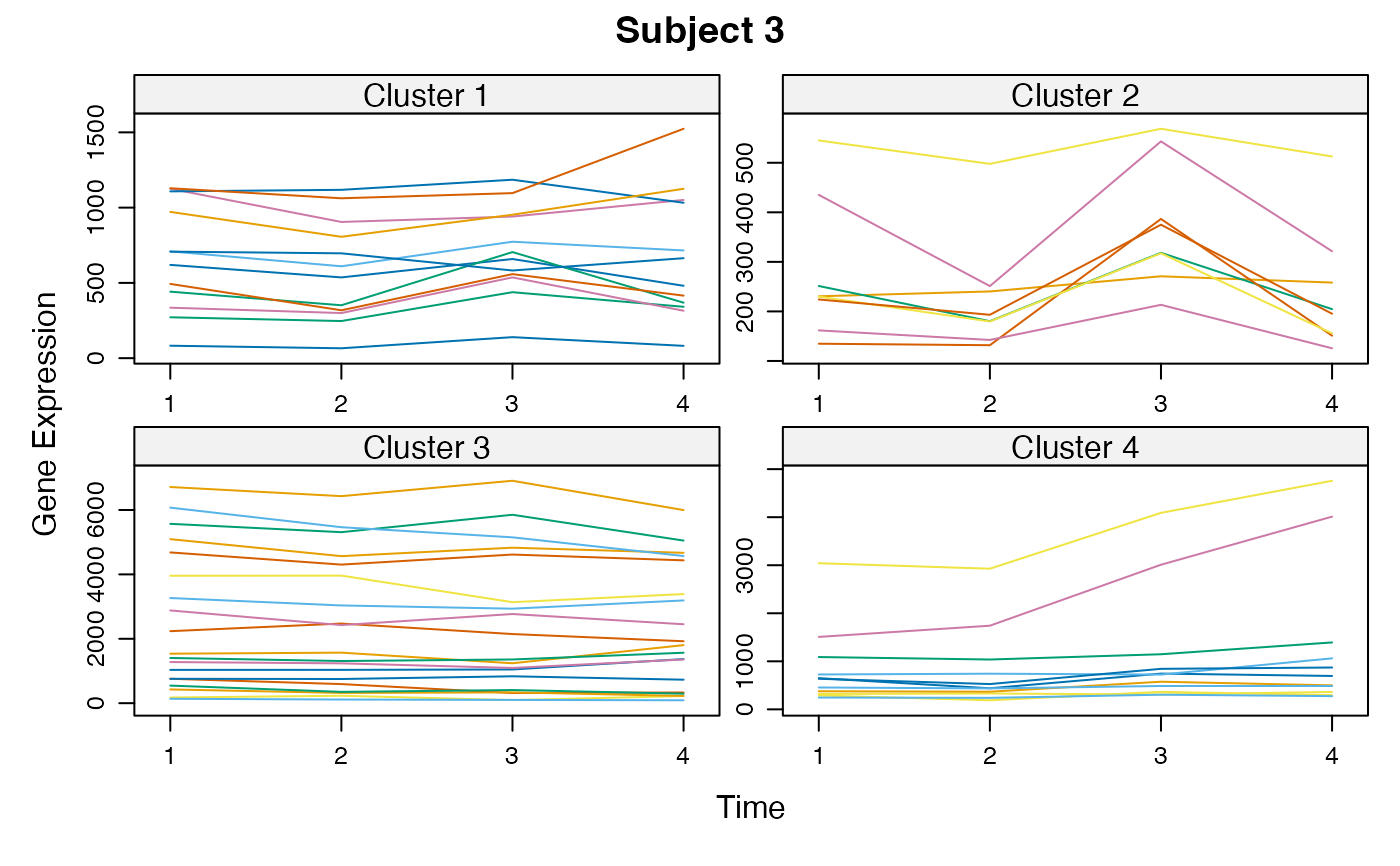
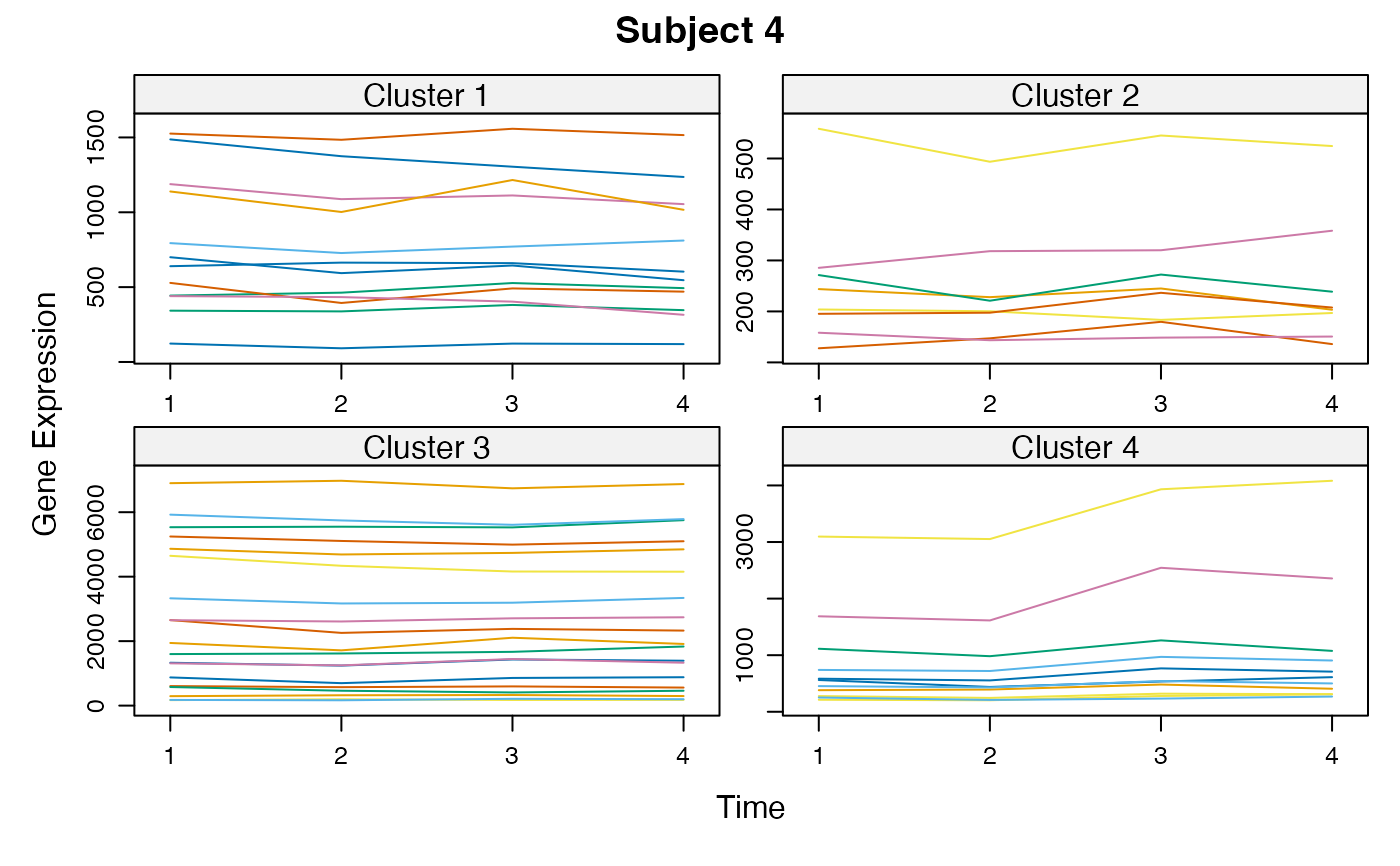
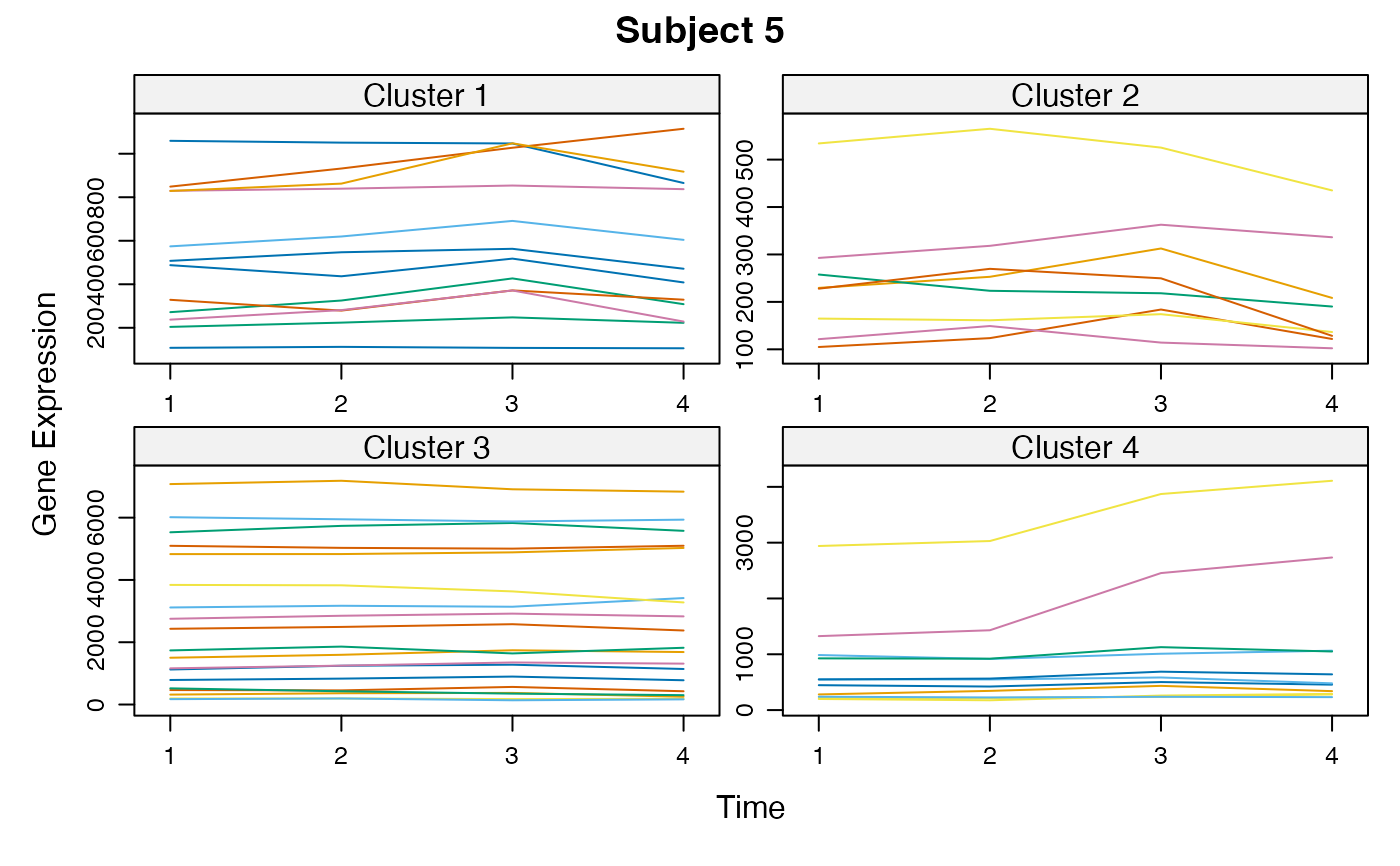
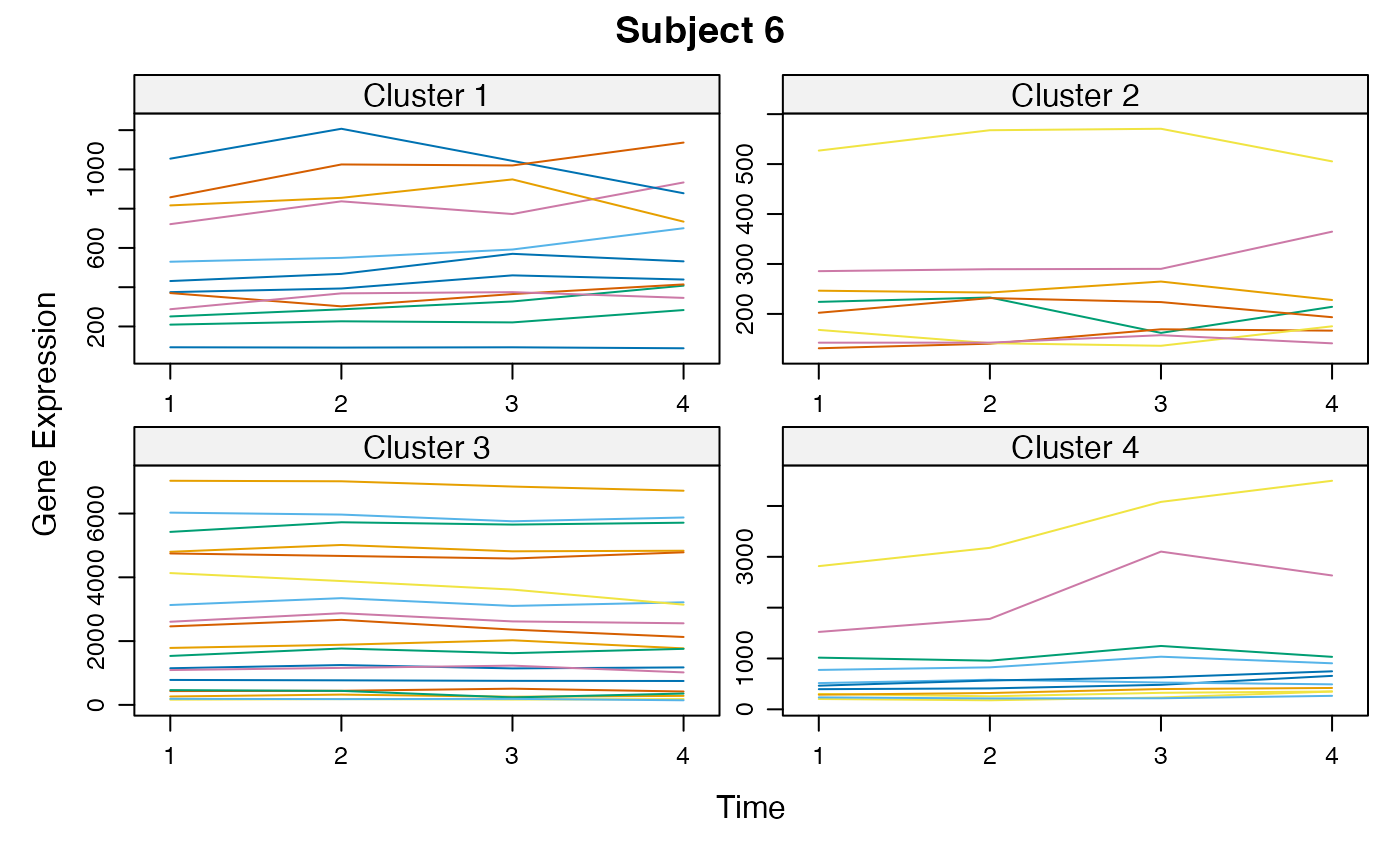
Cluster a omics_array object: performs the clustering.
Source:R/omics_array.R
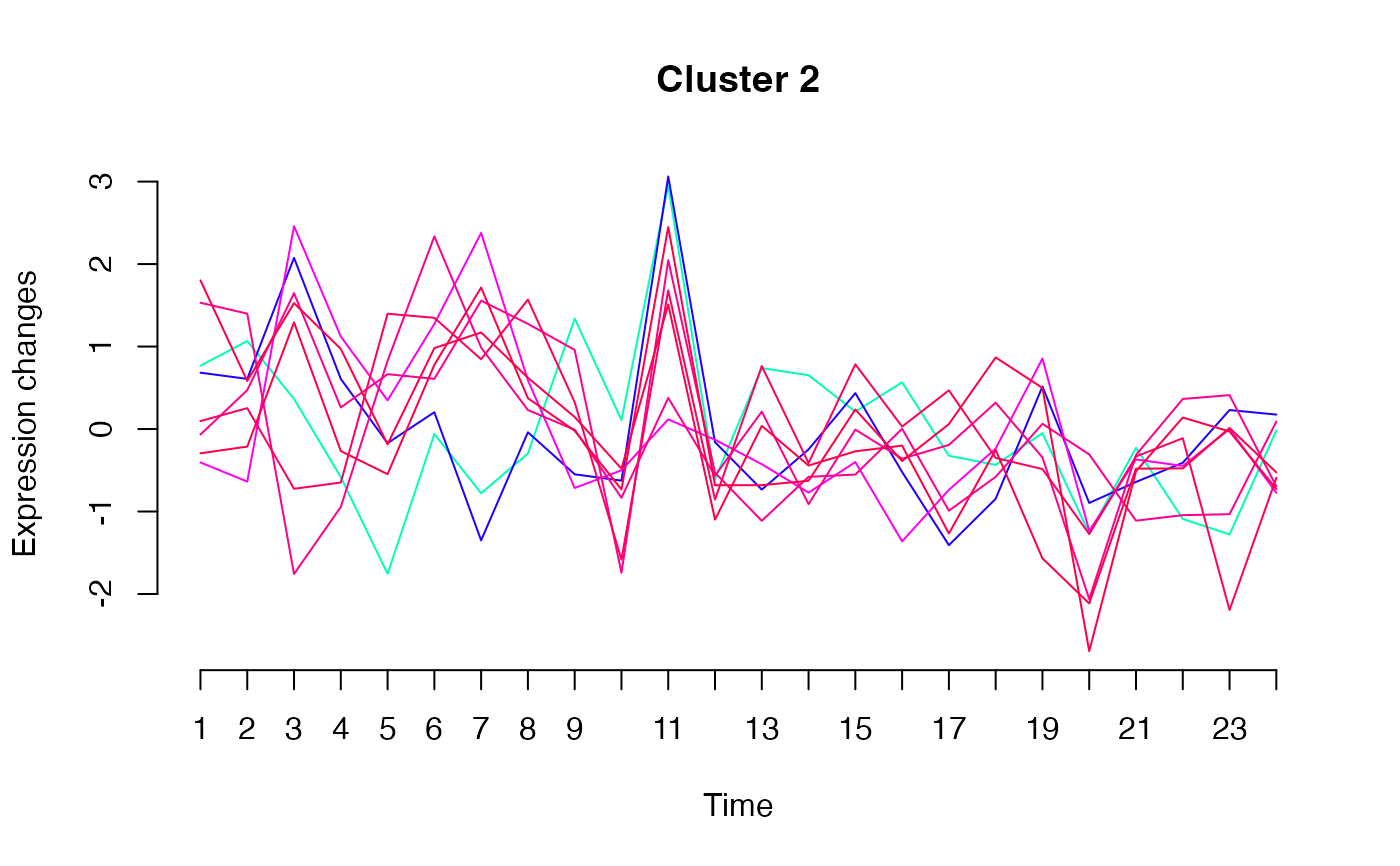
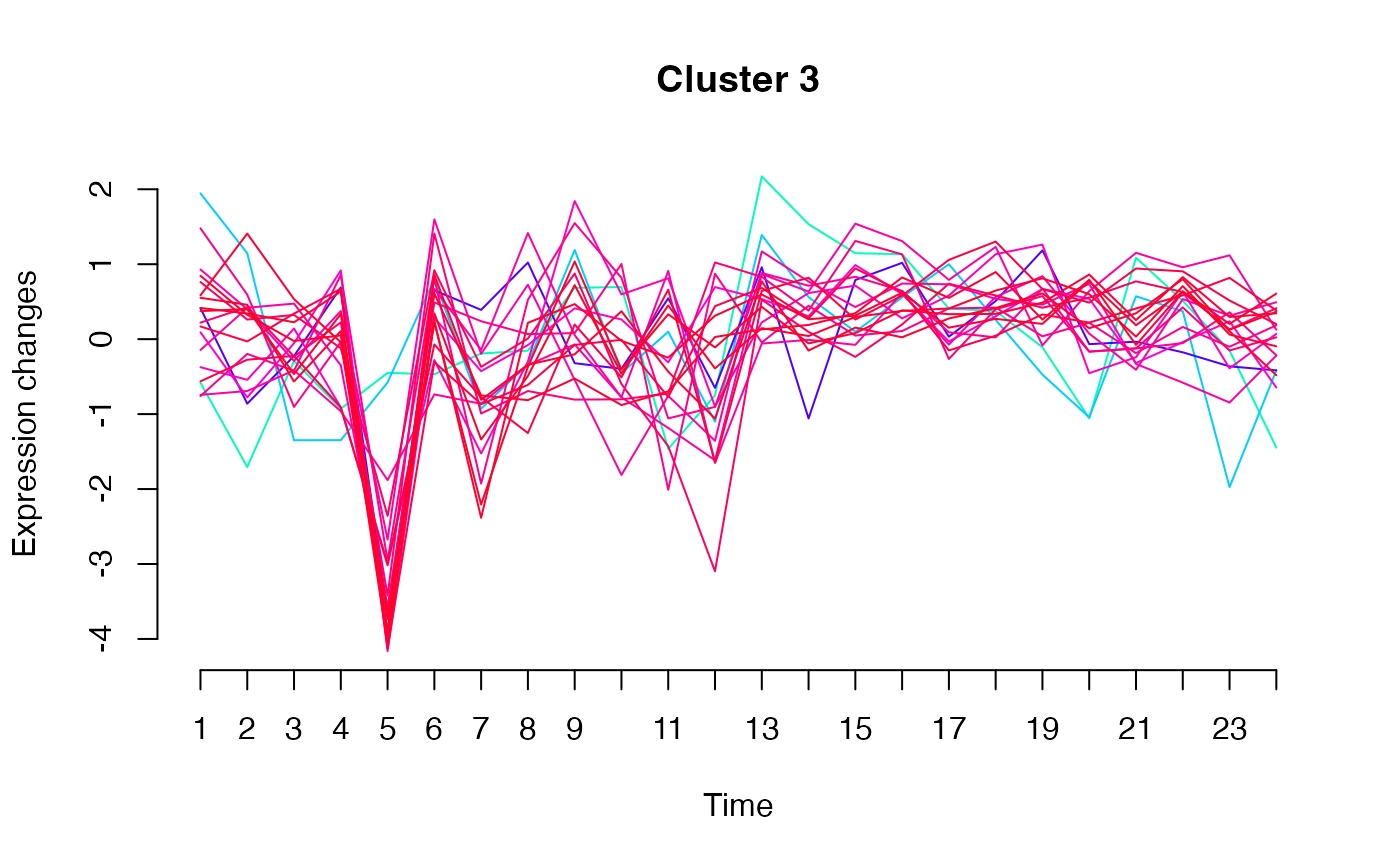
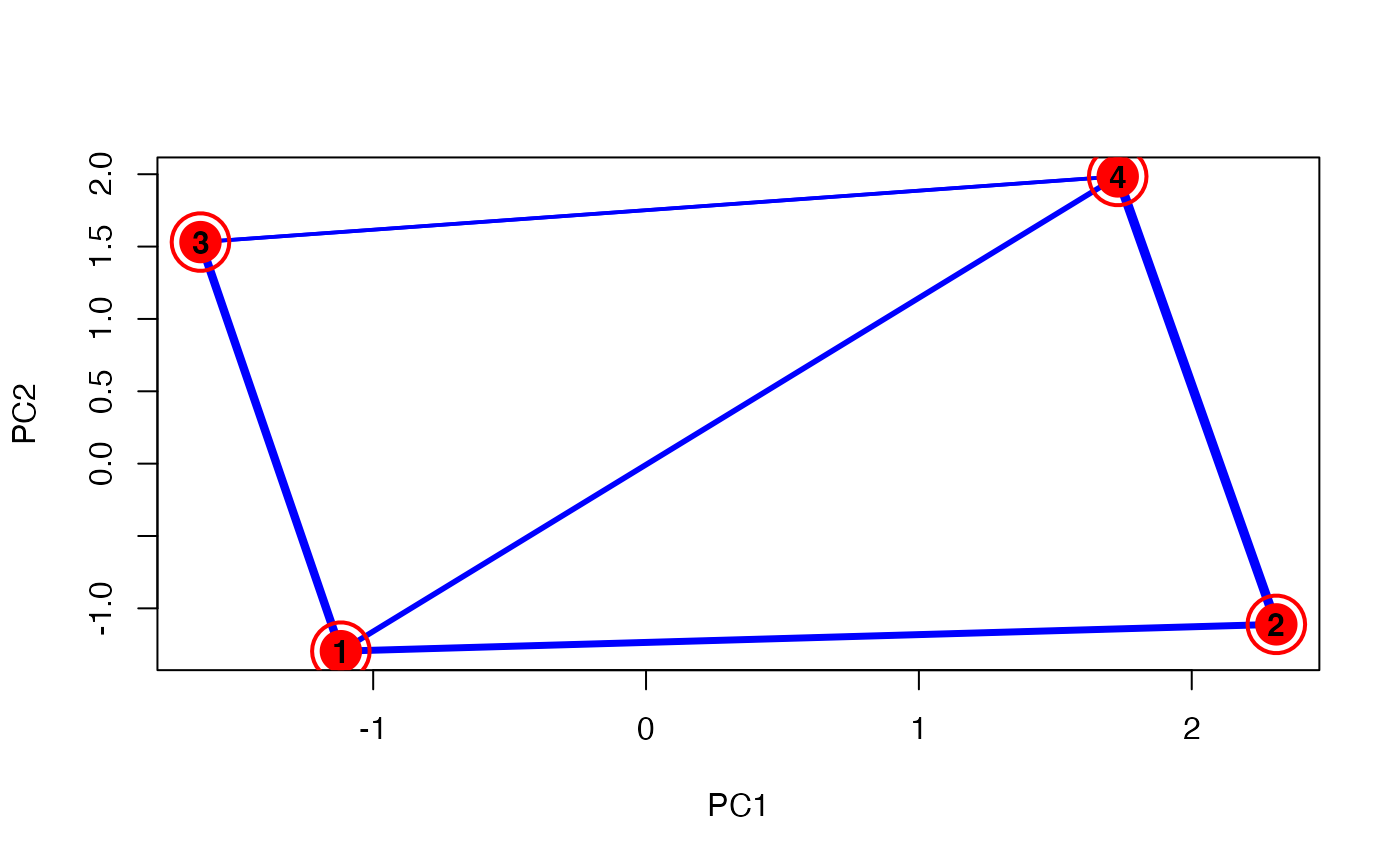
unsupervised_clustering-omics_array-numeric-numeric-method.RdBased on soft clustering performed by the Mfuzz package.
Usage
# S4 method for class 'omics_array,numeric,numeric'
unsupervised_clustering(
M1,
clust,
mestim,
M2 = NULL,
data_log = TRUE,
screen = NULL,
heatmap = TRUE,
new.window = TRUE
)Value
An object of class omics_array with the group slot updated by groups deduced from the soft clustering result.
Examples
if(require(CascadeData)){
data(micro_S, package="CascadeData")
M<-as.omics_array(micro_S[51:100,],1:4,6)
mc<-unsupervised_clustering_auto_m_c(M)
MwithGrp=unsupervised_clustering(M, 4, mc$m, screen=NULL, heatmap=FALSE, new.window = FALSE)
# Other options
unsupervised_clustering(M, 4, mc$m, screen=c(2,2), heatmap=TRUE, new.window = FALSE)
# Plot the clusters
plot(MwithGrp)
}
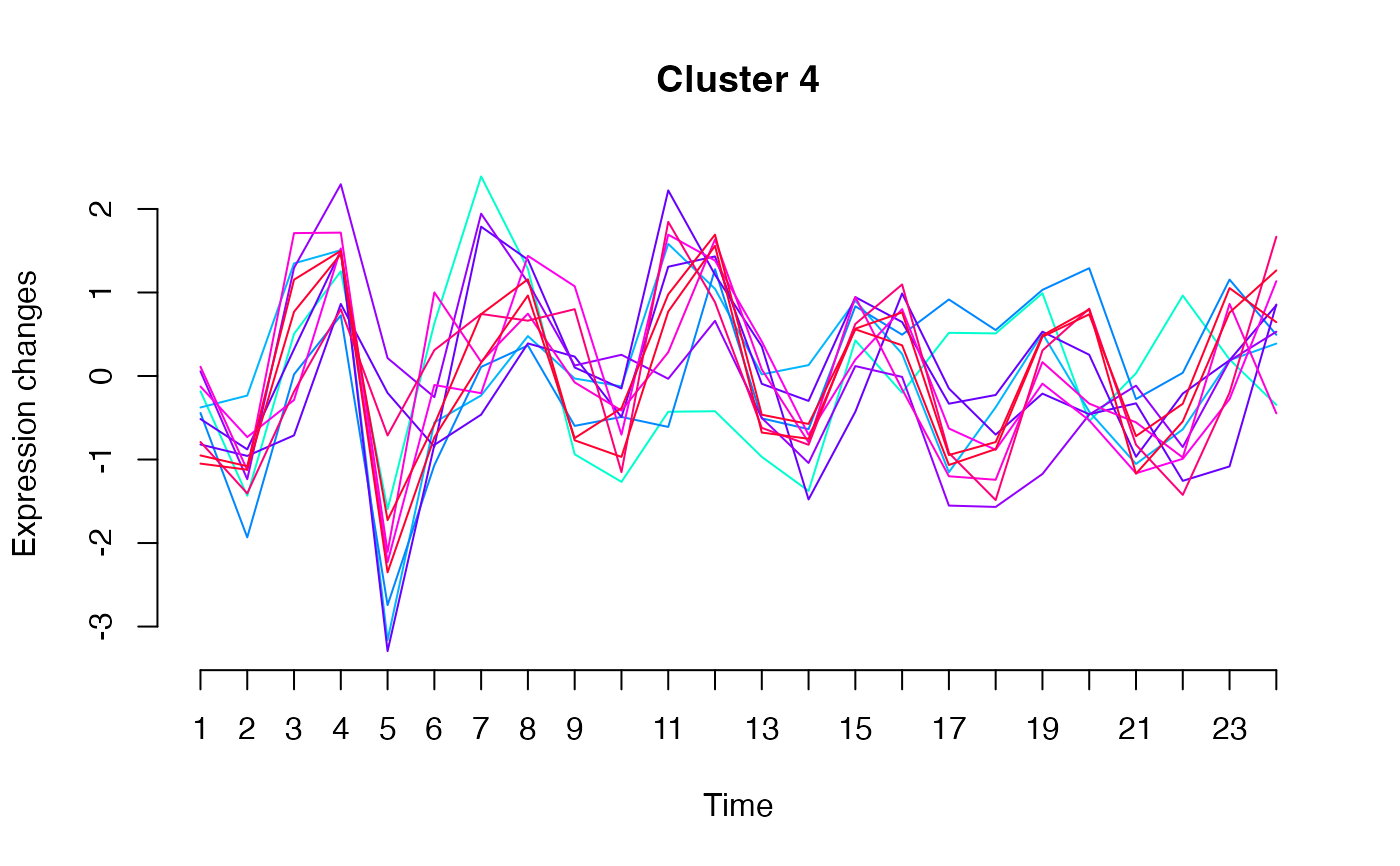
 #>
#> Attaching package: ‘gplots’
#> The following object is masked from ‘package:plotrix’:
#>
#> plotCI
#> The following object is masked from ‘package:stats’:
#>
#> lowess
#>
#> Attaching package: ‘gplots’
#> The following object is masked from ‘package:plotrix’:
#>
#> plotCI
#> The following object is masked from ‘package:stats’:
#>
#> lowess