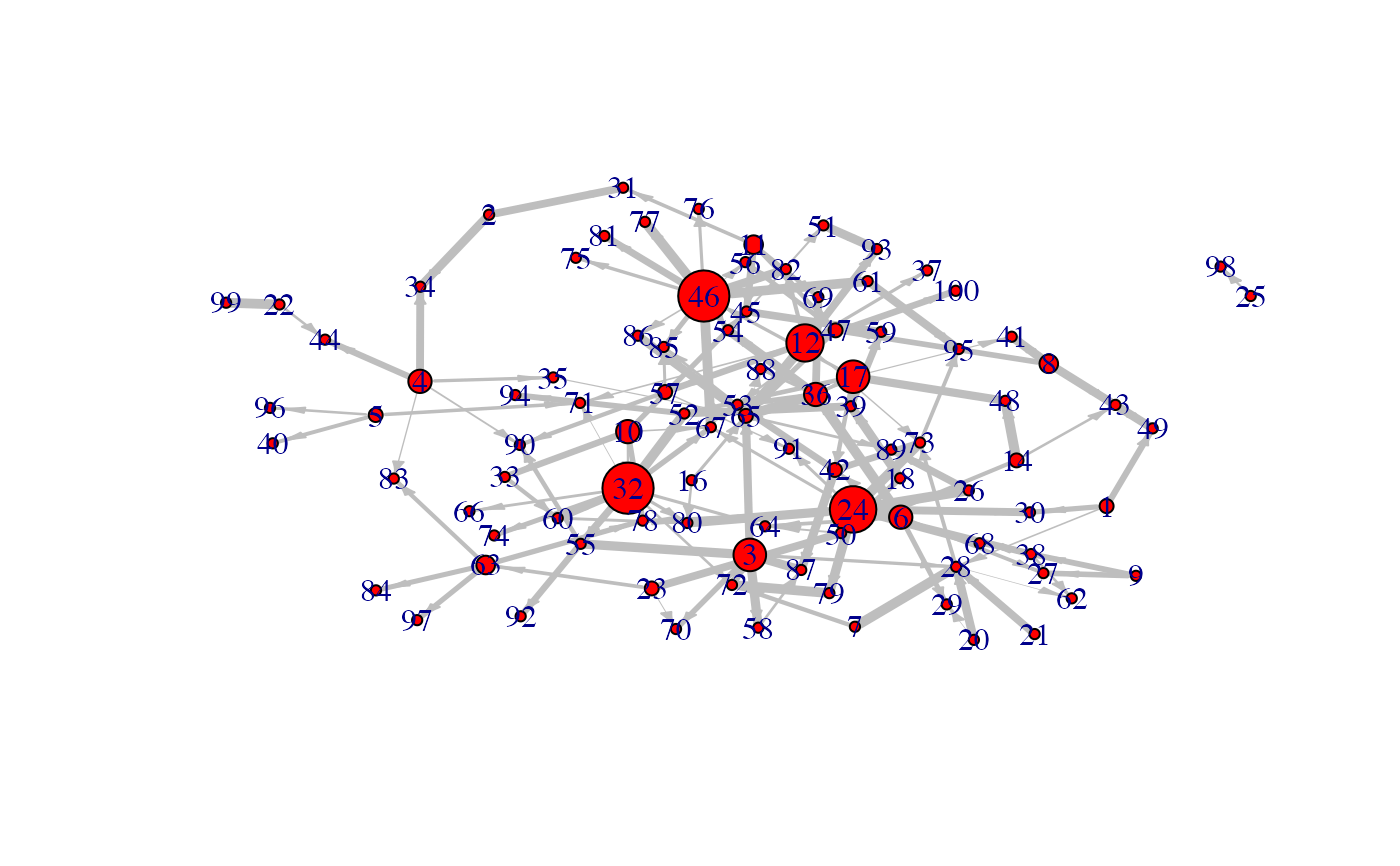
Generates a network.
Arguments
- nb
Integer. The number of genes.
- time_label
Vector. The time points measurements.
- exp
The exponential parameter, as in the barabasi.game function in igraph package.
- init
The attractiveness of the vertices with no adjacent edges. See barabasi.game function.
- regul
A vector mapping each gene with its number of regulators.
- min_expr
Minimum of strength of a non-zero link
- max_expr
Maximum of strength of a non-zero link
- casc.level
...
References
Jung, N., Bertrand, F., Bahram, S., Vallat, L., and Maumy-Bertrand, M. (2014). Cascade: a R-package to study, predict and simulate the diffusion of a signal through a temporal gene network. Bioinformatics, btt705.
Vallat, L., Kemper, C. A., Jung, N., Maumy-Bertrand, M., Bertrand, F., Meyer, N., ... & Bahram, S. (2013). Reverse-engineering the genetic circuitry of a cancer cell with predicted intervention in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences, 110(2), 459-464.