Fast Principal Component Analysis for Big Data with bigPCAcpp
Frédéric Bertrand
Source:vignettes/bigPCAcpp.Rmd
bigPCAcpp.RmdIntroduction
The bigPCAcpp package provides principal component
analysis (PCA) routines that operate directly on bigmemory::big.matrix
objects. This vignette walks through a complete analysis workflow and
compares the results against the reference implementation from base R’s
prcomp()
to demonstrate numerical agreement.
We will use the classic iris measurement data as a
small, in-memory example. Even for larger data sets stored on disk, the
workflow is identical once a big.matrix descriptor has been
created.
Preparing a big.matrix
library(bigmemory)
library(bigPCAcpp)
iris_mat <- as.matrix(iris[, 1:4])
big_iris <- as.big.matrix(iris_mat, type = "double")Every bigPCAcpp entry point accepts the
big.matrix object directly (and, for compatibility, still
works with external pointers via the @address slot),
allowing analyses without copying data into regular R matrices.
Running PCA with bigPCAcpp
big_pca <- pca_bigmatrix(
xpMat = big_iris,
center = TRUE,
scale = TRUE,
ncomp = 4L,
block_size = 128L
)
str(big_pca)
#> List of 10
#> $ sdev : num [1:4] 1.708 0.956 0.383 0.144
#> $ rotation : num [1:4, 1:4] 0.521 -0.269 0.58 0.565 0.377 ...
#> $ center : num [1:4] 5.84 3.06 3.76 1.2
#> $ scale : num [1:4] 0.828 0.436 1.765 0.762
#> $ column_sd : num [1:4] 0.828 0.436 1.765 0.762
#> $ eigenvalues : num [1:4] 2.9185 0.914 0.1468 0.0207
#> $ explained_variance : num [1:4] 0.72962 0.22851 0.03669 0.00518
#> $ cumulative_variance: num [1:4] 0.73 0.958 0.995 1
#> $ covariance : num [1:4, 1:4] 1 -0.118 0.872 0.818 -0.118 ...
#> $ nobs : num 150
#> - attr(*, "backend")= chr "bigmemory"
#> - attr(*, "class")= chr [1:3] "bigpca_bigmemory" "bigpca" "list"The returned list mirrors the structure of a prcomp
object: singular values (sdev), rotation matrix
(rotation), optional centering and scaling vectors, and
additional diagnostics including the covariance matrix and explained
variance ratios.
Comparing against prcomp
base_pca <- prcomp(iris_mat, center = TRUE, scale. = TRUE)
align_columns <- function(reference, target) {
aligned <- target
cols <- min(ncol(reference), ncol(target))
for (j in seq_len(cols)) {
ref <- reference[, j]
tgt <- target[, j]
if (sum((ref - tgt)^2) > sum((ref + tgt)^2)) {
aligned[, j] <- -tgt
}
}
aligned
}
rotation_aligned <- align_columns(base_pca$rotation, big_pca$rotation)
max_rotation_error <- max(abs(rotation_aligned - base_pca$rotation))
max_sdev_error <- max(abs(big_pca$sdev - base_pca$sdev))
big_scores <- pca_scores_bigmatrix(
xpMat = big_iris,
rotation = big_pca$rotation,
center = big_pca$center,
scale = big_pca$scale,
block_size = 128L
)
scores_aligned <- align_columns(base_pca$x, big_scores)
max_score_error <- max(abs(scores_aligned - base_pca$x))
c(
rotation = max_rotation_error,
sdev = max_sdev_error,
scores = max_score_error
)
#> rotation sdev scores
#> 3.441691e-15 1.110223e-15 7.993606e-15The maximum absolute deviations between the base implementation and bigPCAcpp are negligible (on the order of numerical precision), showing that the out-of-core algorithm faithfully reproduces the same components and scores.
Variable relationships
The exported helpers expose common PCA diagnostics without requiring the original data matrix in memory.
loadings <- pca_variable_loadings(big_pca$rotation, big_pca$sdev)
correlations <- pca_variable_correlations(
big_pca$rotation,
big_pca$sdev,
big_pca$column_sd,
big_pca$scale
)
contributions <- pca_variable_contributions(loadings)
head(loadings)
#> [,1] [,2] [,3] [,4]
#> [1,] 0.8901688 0.36082989 0.27565767 -0.03760602
#> [2,] -0.4601427 0.88271627 -0.09361987 0.01777631
#> [3,] 0.9915552 0.02341519 -0.05444699 0.11534978
#> [4,] 0.9649790 0.06399985 -0.24298265 -0.07535950
head(correlations)
#> [,1] [,2] [,3] [,4]
#> [1,] 0.8901688 0.36082989 0.27565767 -0.03760602
#> [2,] -0.4601427 0.88271627 -0.09361987 0.01777631
#> [3,] 0.9915552 0.02341519 -0.05444699 0.11534978
#> [4,] 0.9649790 0.06399985 -0.24298265 -0.07535950
head(contributions)
#> [,1] [,2] [,3] [,4]
#> [1,] 0.27150969 0.1424440565 0.51777574 0.06827052
#> [2,] 0.07254804 0.8524748749 0.05972245 0.01525463
#> [3,] 0.33687936 0.0005998389 0.02019990 0.64232089
#> [4,] 0.31906291 0.0044812296 0.40230191 0.27415396
range(correlations)
#> [1] -0.4601427 0.9915552Loadings match the scaled rotation matrix, correlations honour whether the PCA was computed on standardised variables and remain bounded between -1 and 1, and contributions report the relative importance of each variable within a component.
Visualising PCA results
The companion plotting helpers make it straightforward to inspect the components returned by bigPCAcpp.
pca_plot_scree(big_pca)
Scree plot of variance explained by each component.
The scree plot summarises how much variance each component explains and makes it easy to identify natural cutoffs.
pca_plot_scores(
big_iris,
rotation = big_pca$rotation,
center = big_pca$center,
scale = big_pca$scale,
max_points = nrow(big_iris),
sample = "head"
)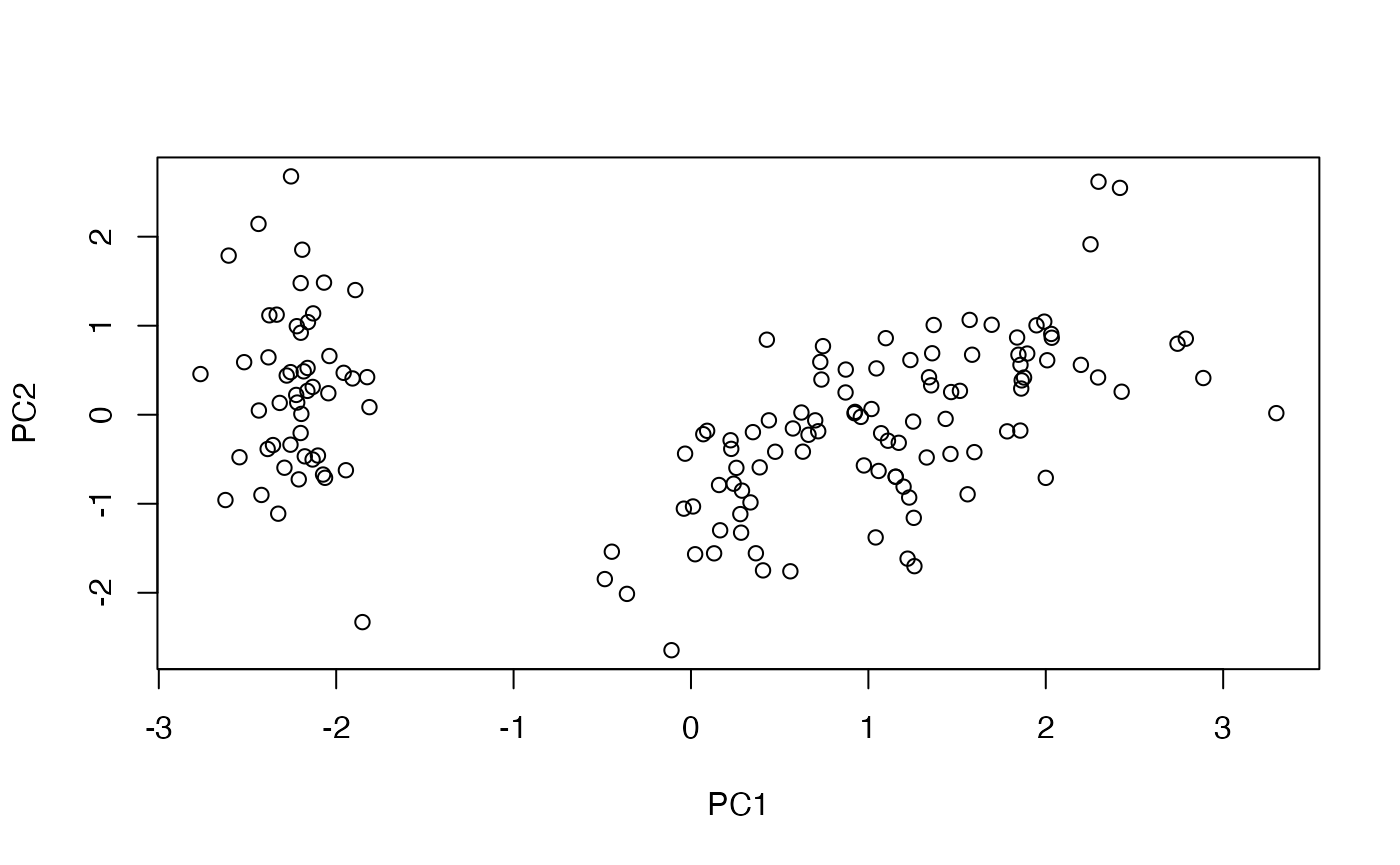
Scores for the first two principal components.
Score plots provide a quick way to compare sample relationships using the requested principal components without materialising the full score matrix.
pca_plot_correlation_circle(
correlations,
components = c(1L, 2L)
)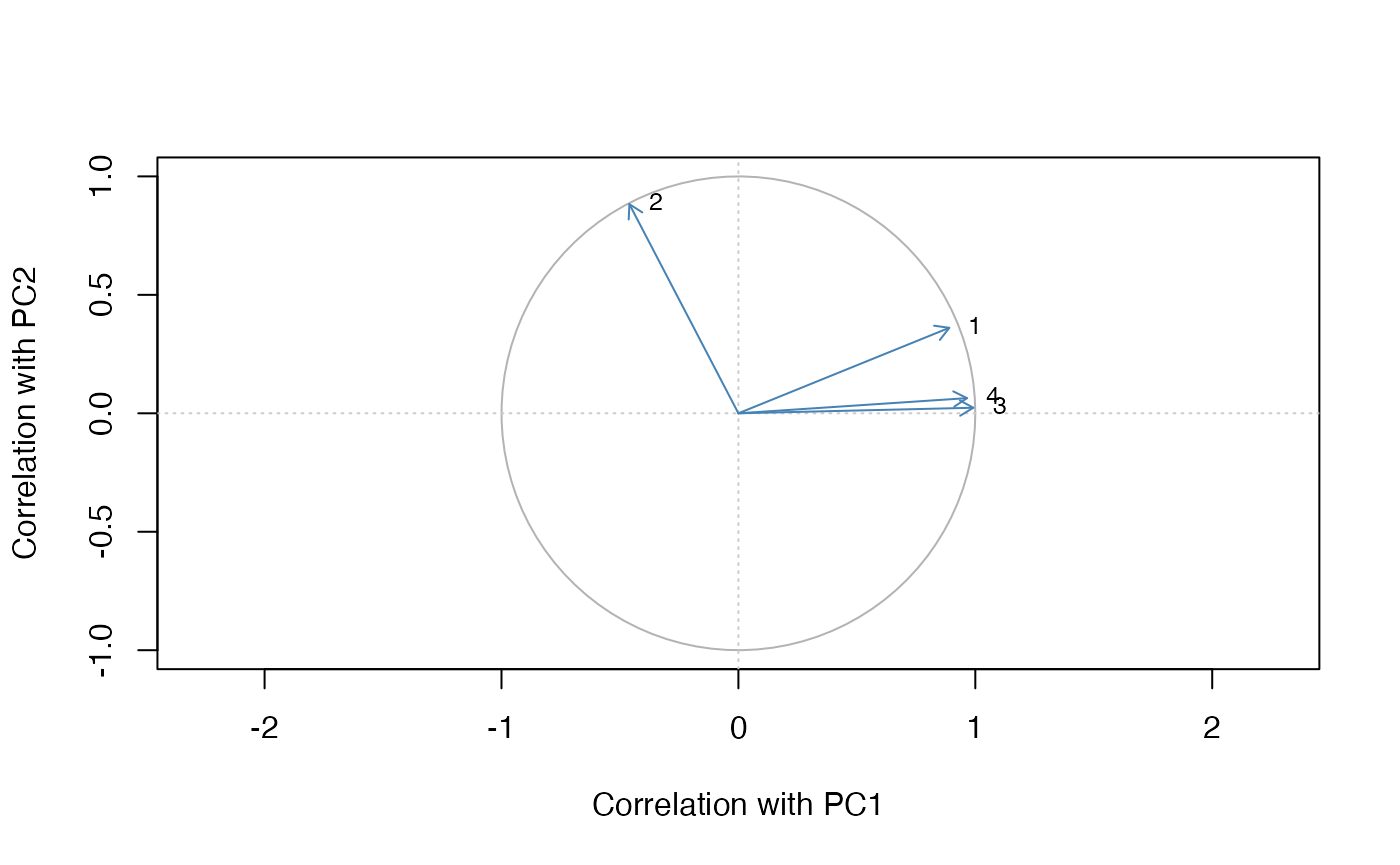
Correlation circle highlighting how variables align with the first two components.
Correlation circles visualise how each variable aligns with the chosen components, making it easy to spot groups of features that contribute in a similar direction.
pca_plot_biplot(
big_scores,
loadings,
components = c(1L, 2L)
)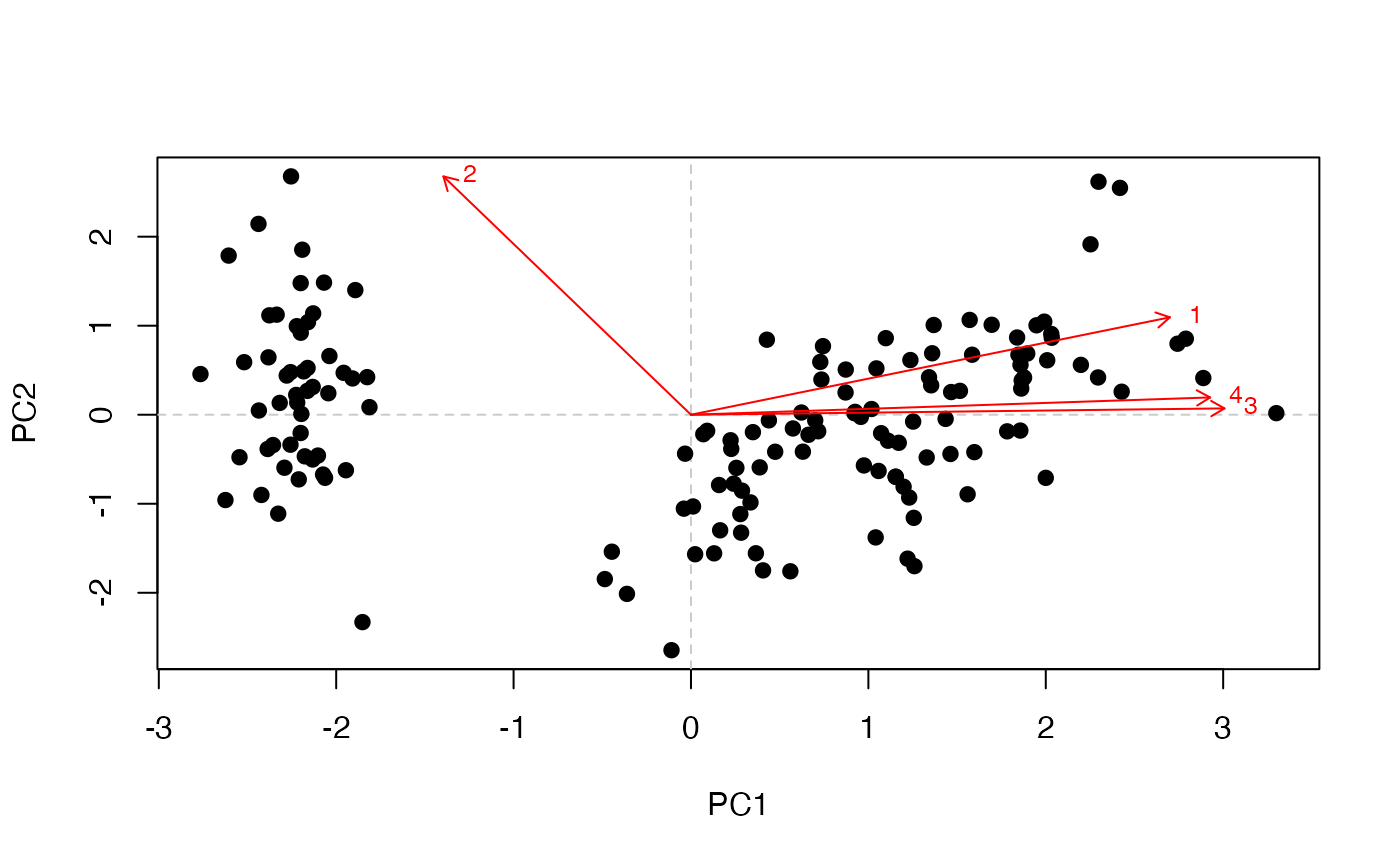
Biplot combining sample scores and variable loadings.
The biplot overlays sample scores with the variable loadings, providing a joint view of how observations cluster along the selected components and which features drive those separations.
Singular value decomposition helpers
The package also exposes building blocks for singular value
decomposition (SVD) that operate directly on big.matrix
instances, which can be useful for custom pipelines that only need the
singular vectors or values.
svd_res <- svd_bigmatrix(big_iris, nu = 2L, nv = 2L, block_size = 128L)
svd_res$d
#> [1] 95.959914 17.761034 3.460931 1.884826The svd_bigmatrix() helper mirrors base R’s svd()
but streams data in manageable blocks, making it practical for large
file-backed matrices.
Robust PCA and SVD
When data contain outliers, the robust variants supplied by bigPCAcpp help stabilise the recovered components by down-weighting leverage points.
robust_pca <- pca_robust(iris_mat, ncomp = 4L)
robust_pca$sdev
#> [1] 1.9153872 0.5075338 0.3074052 0.2357631
robust_pca$robust_weights[1:10]
#> [1] 0.02469728 0.07676234 0.43335242 0.67140174 0.67743015 0.74308617
#> [7] 0.65577512 0.74857591 0.79772190 0.68421285The robust PCA interface accepts regular dense matrices, computes robust estimates of location and scale, and returns component scores along with the per-row weights and iteration counts used by the re-weighted SVD.
The underlying solver is also exposed directly for bespoke workflows that only need a robust singular value decomposition.
robust_svd <- svd_robust(iris_mat, ncomp = 3L)
robust_svd$d
#> [1] 85.31120 16.25951 3.14779
robust_svd$weights[1:10]
#> [1] 1 1 1 1 1 1 1 1 1 1The robust SVD returns singular values, left singular vectors, and the weights assigned to each observation, which can be combined with classical SVD results to assess the influence of individual rows.
Next steps for larger data
For on-disk matrices created with
filebacked.big.matrix(), pass the descriptor pointer to
pca_bigmatrix() and the algorithm will stream data in
blocks, keeping memory usage bounded. Component scores can likewise be
generated in batches using
pca_scores_stream_bigmatrix().
library(bigmemory)
library(bigPCAcpp)
path <- tempfile(fileext = ".bin")
desc <- paste0(path, ".desc")
bm <- filebacked.big.matrix(
nrow = nrow(iris_mat),
ncol = ncol(iris_mat),
type = "double",
backingfile = basename(path),
backingpath = dirname(path),
descriptorfile = basename(desc)
)
bm[,] <- iris_mat
pca <- pca_bigmatrix(bm, center = TRUE, scale = TRUE, ncomp = 4)
scores <- filebacked.big.matrix(
nrow = nrow(bm),
ncol = ncol(pca$rotation),
type = "double",
backingfile = "scores.bin",
backingpath = dirname(path),
descriptorfile = "scores.desc"
)
pca_scores_stream_bigmatrix(
bm,
scores,
pca$rotation,
center = pca$center,
scale = pca$scale
)
#> <pointer: 0x1507a2210>Once the scores are stored on disk, they can be sampled and plotted just like the in-memory workflow:
pca_plot_scores(
bm,
rotation = pca$rotation,
center = pca$center,
scale = pca$scale,
components = c(1L, 2L),
max_points = nrow(bm),
sample = "head"
)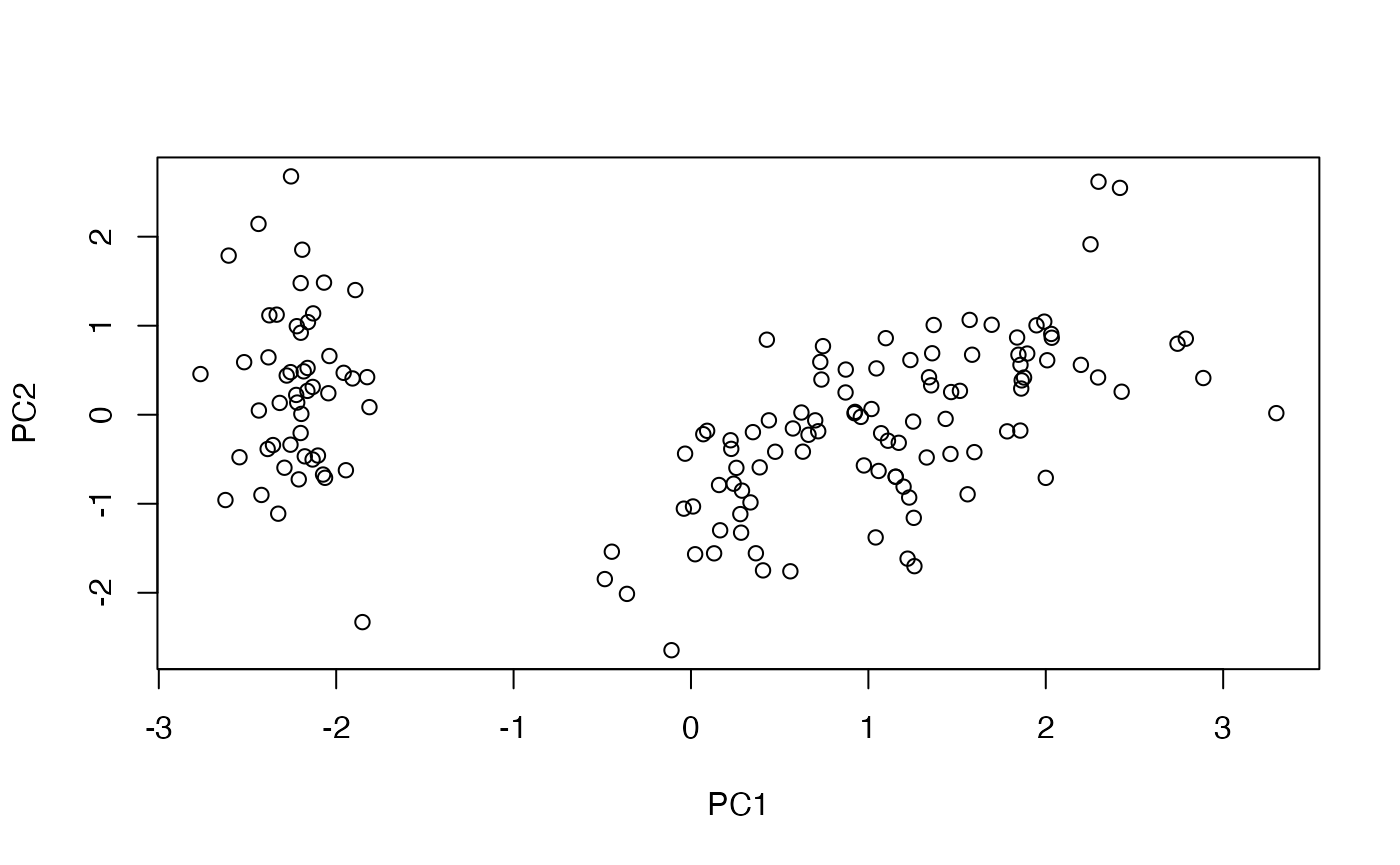
Scores streamed from a file-backed big.matrix.
library(bigmemory)
library(bigPCAcpp)
path <- tempfile(fileext = ".bin")
desc <- paste0(path, ".desc")
bm <- filebacked.big.matrix(
nrow = 5000,
ncol = 50,
type = "double",
backingfile = basename(path),
backingpath = dirname(path),
descriptorfile = basename(desc)
)
pca <- pca_bigmatrix(bm, center = TRUE, scale = TRUE, ncomp = 5)
scores <- filebacked.big.matrix(
nrow = nrow(bm),
ncol = ncol(pca$rotation),
type = "double",
backingfile = "scores.bin",
backingpath = dirname(path),
descriptorfile = "scores.desc"
)
pca_scores_stream_bigmatrix(
bm,
scores,
pca$rotation,
center = pca$center,
scale = pca$scale
)With these building blocks, bigPCAcpp enables analyses that match the accuracy of in-memory PCA workflows while scaling to data sets that exceed RAM.