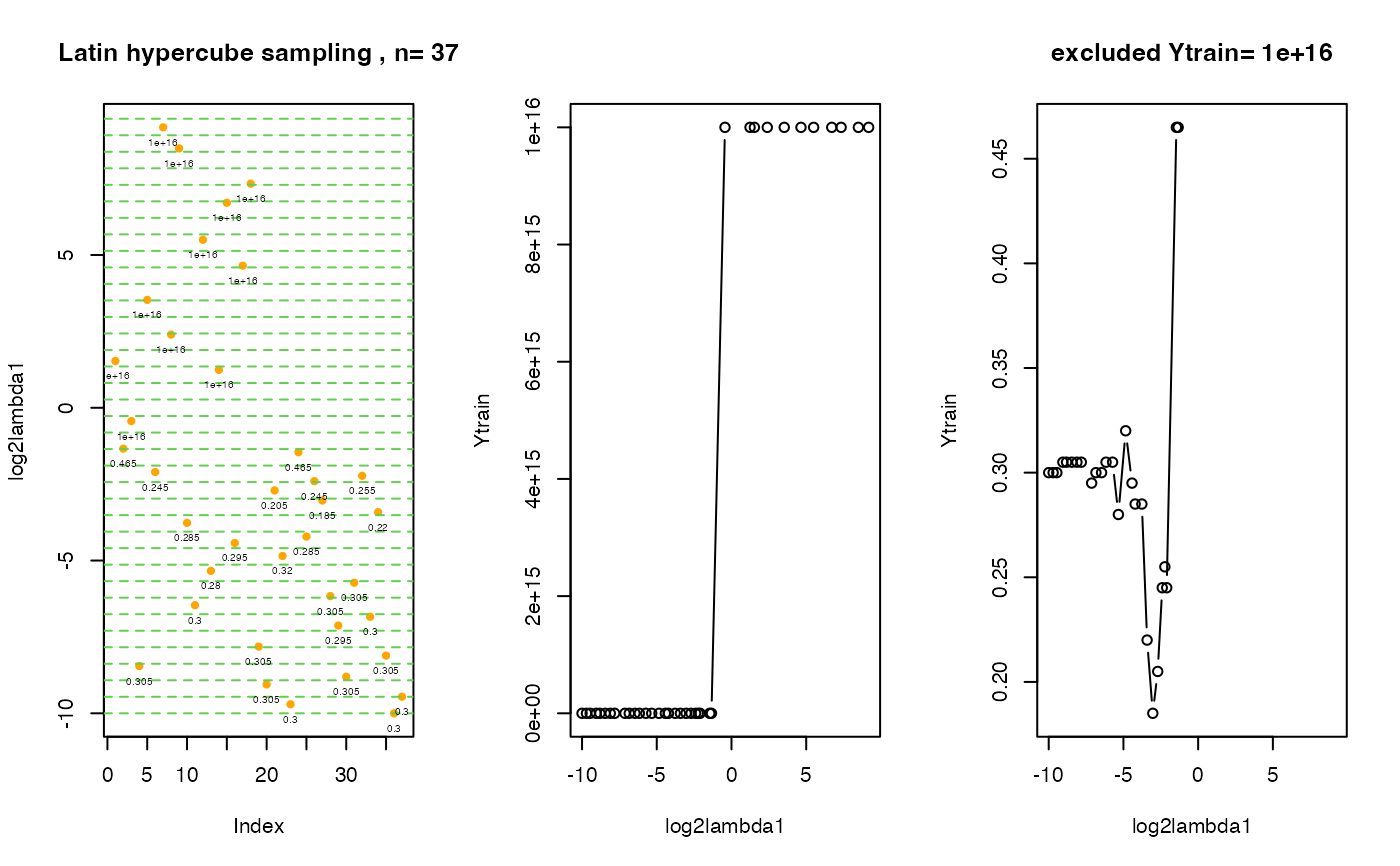
Plot Interval Search Plot Visited Points and the Q Values.
plot.EPSGO.parms.RdFor interval search plot visited points and the Q values (=Ytrain) exclude: for D=1 make an additional plot: skip values for empty model, for example: Ytrain.exclude=10^16.
Author
Natalia Becker
natalie_becker@gmx.de
References
Froehlich, H. and Zell, A. (2005) "Effcient parameter selection for support vector machines in classification and regression via model-based global optimization" In Proc. Int. Joint Conf. Neural Networks, 1431-1438 .
Examples
# \donttest{
seed <- 123
train<-sim.data(n = 200, ng = 100, nsg = 10, corr=FALSE, seed=seed )
print(str(train))
#> List of 3
#> $ x : num [1:100, 1:200] 0.547 0.635 -0.894 0.786 2.028 ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : chr [1:100] "pos1" "pos2" "pos3" "pos4" ...
#> .. ..$ : chr [1:200] "1" "2" "3" "4" ...
#> $ y : Named num [1:200] 1 -1 -1 1 -1 -1 1 -1 -1 -1 ...
#> ..- attr(*, "names")= chr [1:200] "1" "2" "3" "4" ...
#> $ seed: num 123
#> NULL
Q.func<- ".calc.scad"
bounds=t(data.frame(log2lambda1=c(-10, 10)))
colnames(bounds)<-c("lower", "upper")
print("start interval search")
#> [1] "start interval search"
# computation intensive;
# for demostration reasons only for the first 100 features
# and only for 10 iterations maxIter=10, default maxIter=700
system.time(fit<-EPSGO(Q.func, bounds=bounds, parms.coding="log2", fminlower=0,
show='none', N=21, maxevals=500,
pdf.name=NULL, seed=seed,
verbose=FALSE,
# Q.func specific parameters:
x.svm=t(train$x)[,1:100], y.svm=train$y,
inner.val.method="cv",
cross.inner=5, maxIter=10 ))
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> Warning: Coercing LHS to a list
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> ...done
#> user system elapsed
#> 6.042 0.125 6.199
print(paste("minimal 5-fold cv error:", fit$fmin, "by log2(lambda1)=", fit$xmin))
#> [1] "minimal 5-fold cv error: 0.185 by log2(lambda1)= -3.03520543316445"
print(" all lambdas with the same minimum? ")
#> [1] " all lambdas with the same minimum? "
print(fit$ points.fmin)
#> log2lambda1 f
#> 27 -3.035205 0.185
print(paste(fit$neval, "visited points"))
#> [1] "37 visited points"
print(" overview: over all visitied points in tuning parameter space
with corresponding cv errors")
#> [1] " overview: over all visitied points in tuning parameter space \n\t\t\t\twith corresponding cv errors"
print(data.frame(Xtrain=fit$Xtrain, cv.error=fit$Ytrain))
#> log2lambda1 cv.error
#> 1 1.5315485 1.00e+16
#> 2 -1.3383552 4.65e-01
#> 3 -0.4348424 1.00e+16
#> 4 -8.4531580 3.05e-01
#> 5 3.5310332 1.00e+16
#> 6 -2.1035852 2.45e-01
#> 7 9.1782694 1.00e+16
#> 8 2.3956101 1.00e+16
#> 9 8.4905937 1.00e+16
#> 10 -3.7667024 2.85e-01
#> 11 -6.4618650 3.00e-01
#> 12 5.4976793 1.00e+16
#> 13 -5.3380199 2.80e-01
#> 14 1.2415551 1.00e+16
#> 15 6.7099483 1.00e+16
#> 16 -4.4249049 2.95e-01
#> 17 4.6509131 1.00e+16
#> 18 7.3355014 1.00e+16
#> 19 -7.8173000 3.05e-01
#> 20 -9.0531639 3.05e-01
#> 21 -2.7058843 2.05e-01
#> 22 -4.8506733 3.20e-01
#> 23 -9.7038186 3.00e-01
#> 24 -1.4557824 4.65e-01
#> 25 -4.2158845 2.85e-01
#> 26 -2.4017091 2.45e-01
#> 27 -3.0352054 1.85e-01
#> 28 -6.1636759 3.05e-01
#> 29 -7.1268118 2.95e-01
#> 30 -8.8040755 3.05e-01
#> 31 -5.7266564 3.05e-01
#> 32 -2.2278129 2.55e-01
#> 33 -6.8407343 3.00e-01
#> 34 -3.4158114 2.20e-01
#> 35 -8.1115934 3.05e-01
#> 36 -9.9999979 3.00e-01
#> 37 -9.4561642 3.00e-01
# create 3 plots om one screen:
# 1st plot: distribution of initial points in tuning parameter space
# 2nd plot: visited lambda points vs. cv errors
# 3rd plot: the same as the 2nd plot, Ytrain.exclude points are excluded.
# The value cv.error = 10^16 stays for the cv error for an empty model !
.plot.EPSGO.parms (fit$Xtrain, fit$Ytrain,bound=bounds,
Ytrain.exclude=10^16, plot.name=NULL )
 # } # end of \donttest
# } # end of \donttest