Comparing SelectBoost-GAMLSS Variants
Frédéric Bertrand
Cedric, Cnam, Parisfrederic.bertrand@lecnam.net
2025-10-27
Source:vignettes/comparison.Rmd
comparison.RmdThis vignette compares: plain sb_gamlss, the
SelectBoost-integrated SelectBoost_gamlss, grid-based
sb_gamlss_c0_grid + autoboost_gamlss, and the
lightweight fastboost_gamlss.
What you’ll learn
- How the base
sb_gamlss()call differs fromSelectBoost_gamlss()(automatic grouping). - How to sweep c0 values with
sb_gamlss_c0_grid()and letautoboost_gamlss()pick a favourite. - How to generate rapid previews with
fastboost_gamlss()before launching a large bootstrap campaign.
library(gamlss)
#> Loading required package: splines
#> Loading required package: gamlss.data
#>
#> Attaching package: 'gamlss.data'
#> The following object is masked from 'package:datasets':
#>
#> sleep
#> Loading required package: gamlss.dist
#> Loading required package: nlme
#> Loading required package: parallel
#> ********** GAMLSS Version 5.5-0 **********
#> For more on GAMLSS look at https://www.gamlss.com/
#> Type gamlssNews() to see new features/changes/bug fixes.
library(SelectBoost.gamlss)
set.seed(1)
n <- 500
x1 <- rnorm(n); x2 <- rnorm(n); x3 <- rnorm(n); x4 <- rnorm(n)
mu <- 0.5 + 1.2*x1 - 0.8*x3
y <- gamlss.dist::rNO(n, mu = mu, sigma = 1)
dat <- data.frame(y, x1, x2, x3, x4)
# Baseline
b0 <- sb_gamlss(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ x1 + x2 + x3 + x4, sigma_scope = ~ x1 + x2,
B = 50, pi_thr = 0.6, pre_standardize = TRUE, trace = FALSE
)
plot_sb_gamlss(b0)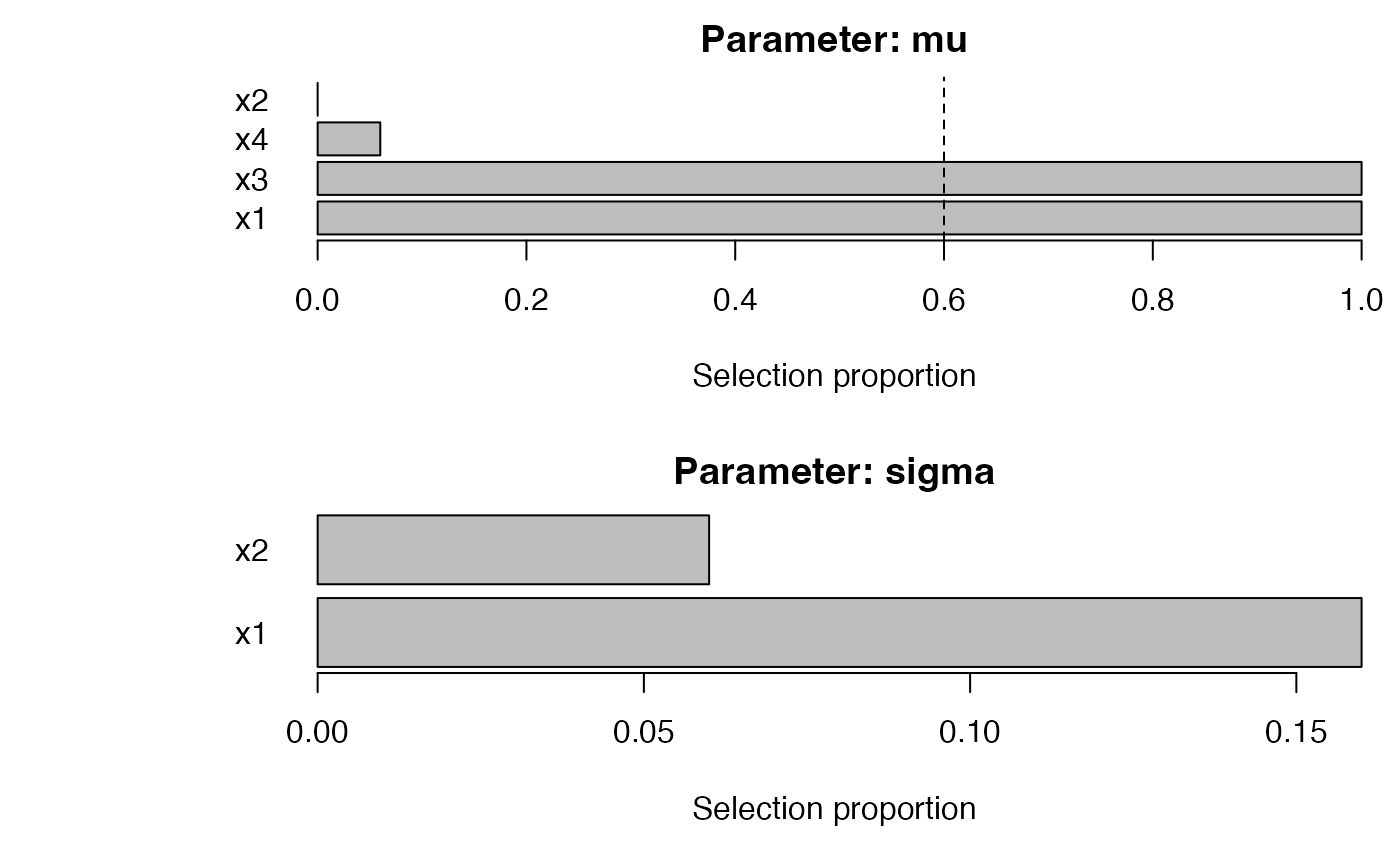
# SelectBoost integration (single c0)
sb <- SelectBoost_gamlss(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ x1 + x2 + x3 + x4, sigma_scope = ~ x1 + x2,
B = 50, pi_thr = 0.6, c0 = 0.5, pre_standardize = TRUE, trace = FALSE
)
summary(sb) |> plot()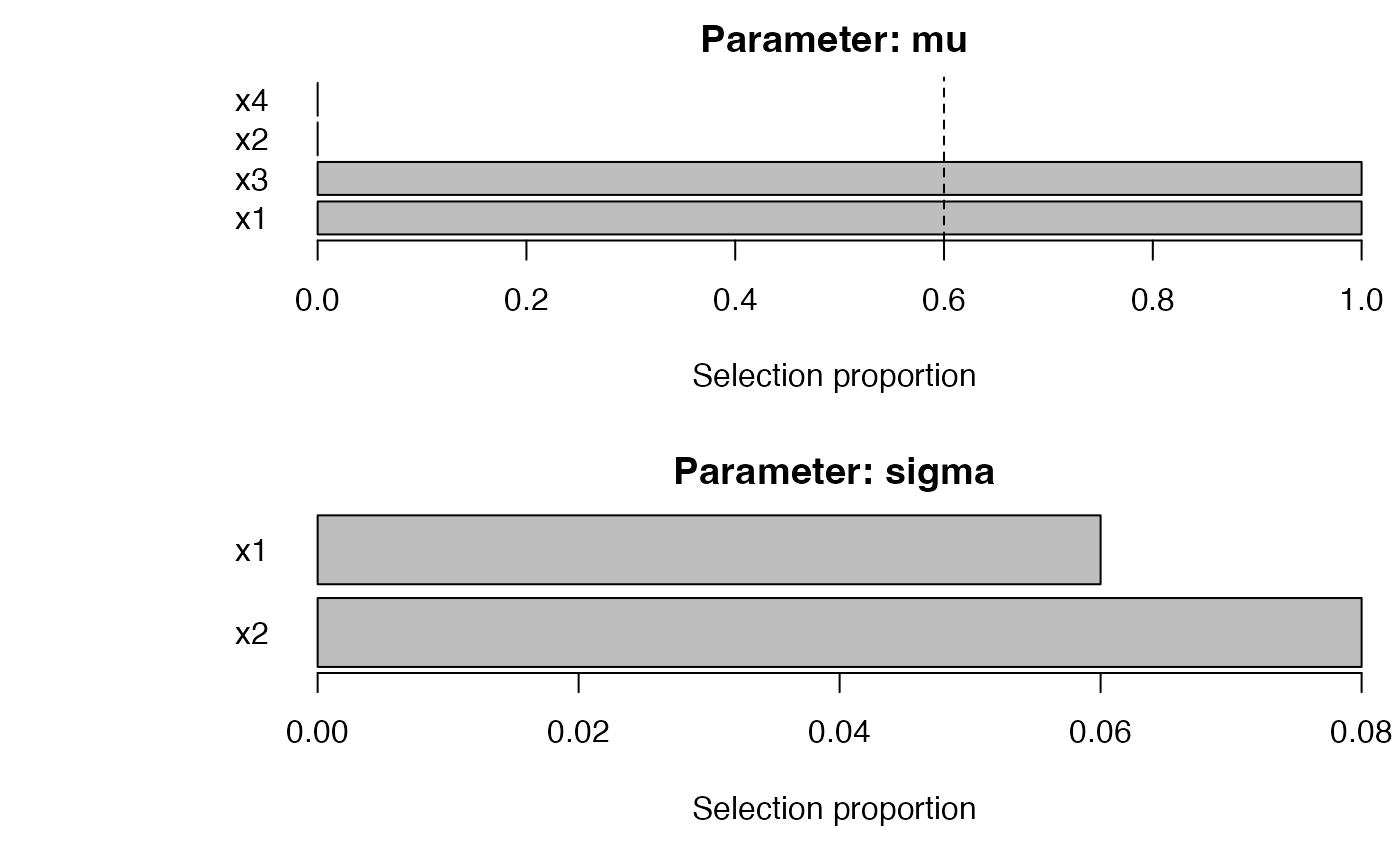
# c0 grid + autoboost
g <- sb_gamlss_c0_grid(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ x1 + x2 + x3 + x4, sigma_scope = ~ x1 + x2,
c0_grid = seq(0.2, 0.8, by = 0.2), B = 40, pi_thr = 0.6, pre_standardize = TRUE, trace = FALSE
)
#> | | | 0% | |================== | 25% | |=================================== | 50% | |==================================================== | 75% | |======================================================================| 100%
plot(g)
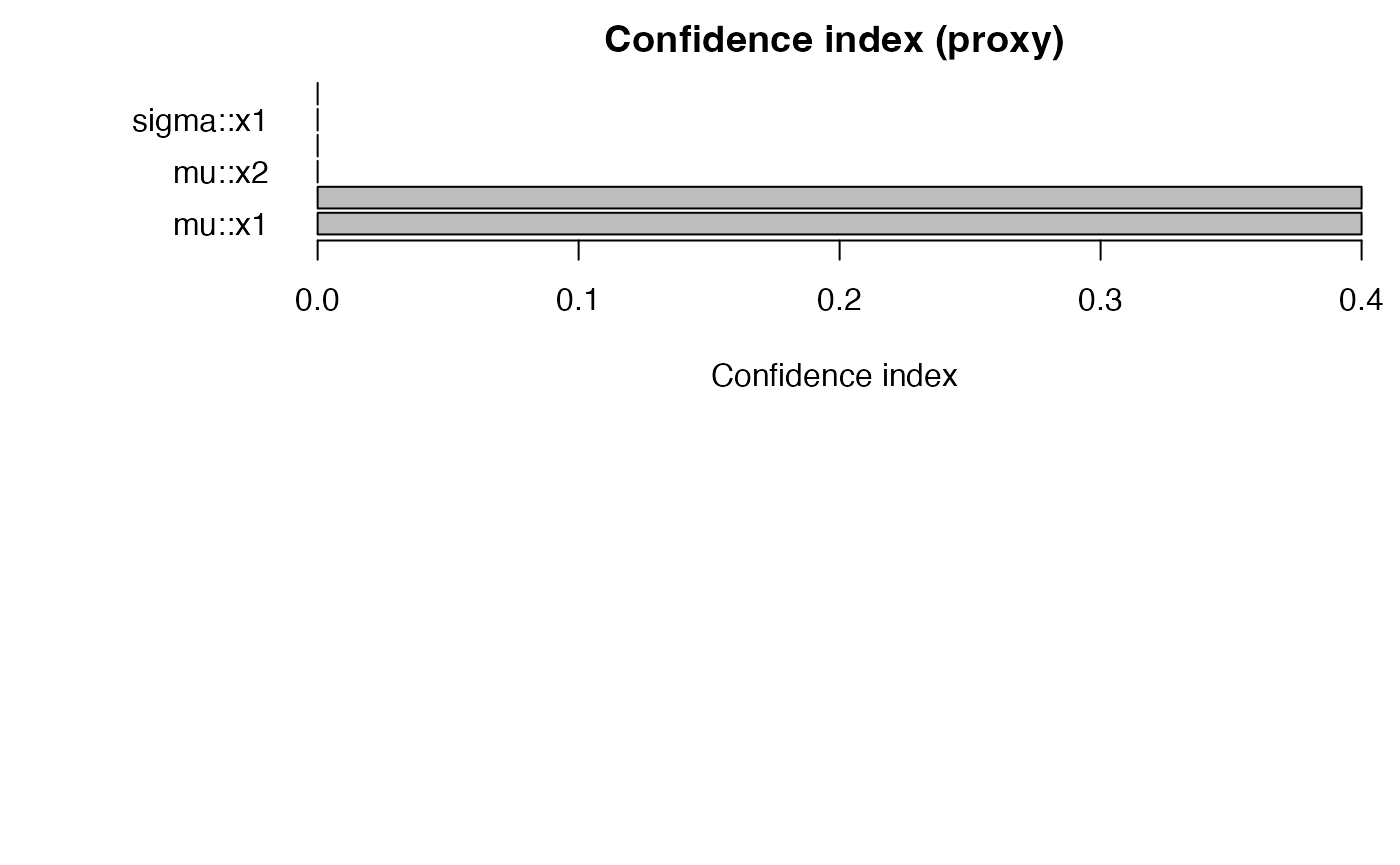
confidence_table(g) |> head()
#> parameter term conf_index cover
#> 1 mu x1 0.4 1
#> 5 mu x3 0.4 1
#> 2 sigma x1 0.0 0
#> 3 mu x2 0.0 0
#> 4 sigma x2 0.0 0
#> 6 mu x4 0.0 0
ab <- autoboost_gamlss(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ x1 + x2 + x3 + x4, sigma_scope = ~ x1 + x2,
c0_grid = seq(0.2, 0.8, by = 0.2), B = 40, pi_thr = 0.6, pre_standardize = TRUE, trace = FALSE
)
#> | | | 0% | |================== | 25% | |=================================== | 50%
#> Warning in RS(): Algorithm RS has not yet converged
#> | |==================================================== | 75% | |======================================================================| 100%
attr(ab, "chosen_c0")
#> [1] 0.2
plot_sb_gamlss(ab)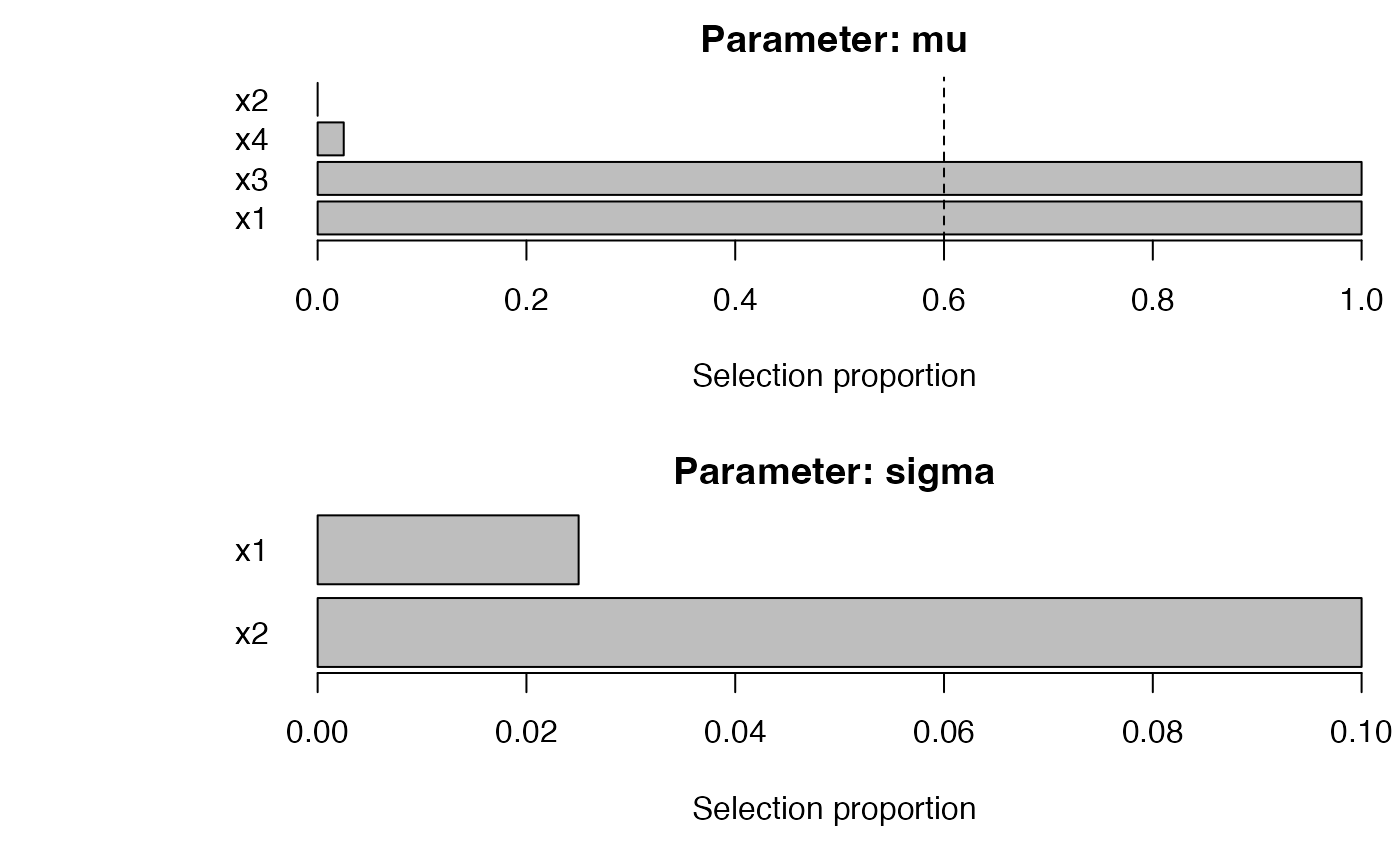
# fastboost (lightweight)
fb <- fastboost_gamlss(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ x1 + x2 + x3 + x4, sigma_scope = ~ x1 + x2,
B = 30, sample_fraction = 0.6, pi_thr = 0.6, pre_standardize = TRUE, trace = FALSE
)
plot_sb_gamlss(fb)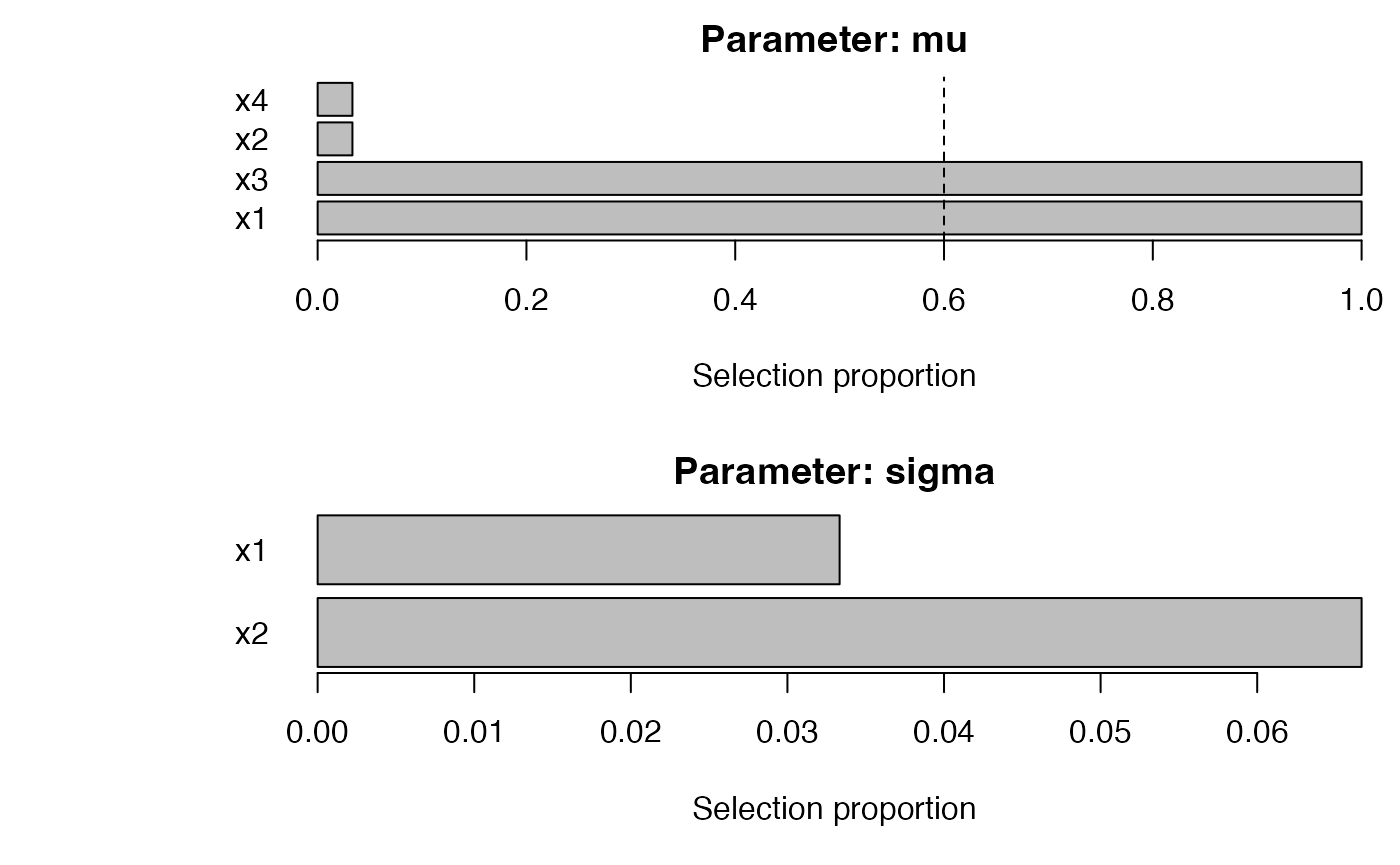
Confidence Functionals (AUSC, Coverage, Quantiles, Weighted, Conservative)
We summarize the stability curves into single-number scores:
-
AUSC: normalized area under
across the
c0_grid. - Thresholded AUSC ( area): only counts confidence above the stability threshold.
-
Coverage: fraction of
c0where . - Quantiles (median/80th/90th): robust distributional summaries of stability.
- Weighted AUSC: apply a weight function to emphasize parts of the grid.
- Conservative AUSC: use Wilson lower bounds for before computing AUSC.
# Using the grid g created above
cf <- confidence_functionals(
g, pi_thr = 0.6,
q = c(0.5, 0.8, 0.9),
weight_fun = function(c0) (1 - c0)^2, # emphasize smaller c0
conservative = TRUE, B = 40, # use lower bounds
method = "trapezoid"
)
head(cf)
#> area area_pos w_area cover sup inf q50
#> 1 0.912375461 0.3123755 0.912375461 1 0.91237546 0.91237546 0.912375461
#> 3 0.912375461 0.3123755 0.912375461 1 0.91237546 0.91237546 0.912375461
#> 6 0.048498421 0.0000000 0.063195402 0 0.07061092 0.00000000 0.055094902
#> 5 0.037500329 0.0000000 0.037371182 0 0.05459420 0.02583556 0.039578888
#> 2 0.006910189 0.0000000 0.002892637 0 0.01382038 0.00000000 0.006910189
#> 4 0.000000000 0.0000000 0.000000000 0 0.00000000 0.00000000 0.000000000
#> q80 q90 parameter term rank_score
#> 1 0.91237546 0.91237546 mu x1 0.638662823
#> 3 0.91237546 0.91237546 mu x3 0.638662823
#> 6 0.07061092 0.07061092 sigma x2 0.014122183
#> 5 0.04558501 0.05008960 sigma x1 0.010017921
#> 2 0.01382038 0.01382038 mu x2 0.002764075
#> 4 0.00000000 0.00000000 mu x4 0.000000000
plot(cf, top = 12, label_top = 8)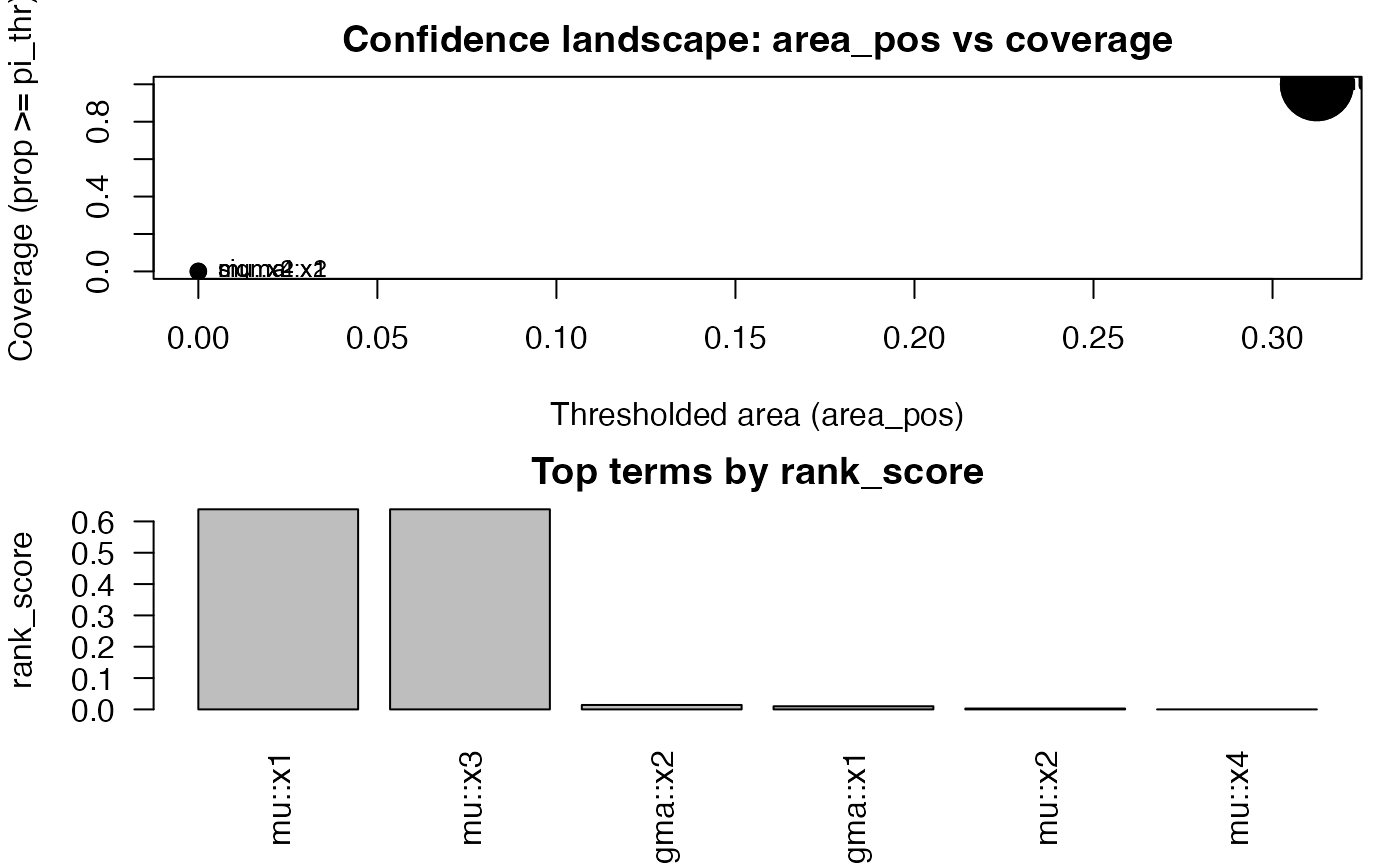
# Inspect stability curves of top terms
top_terms <- head(cf[order(cf$rank_score, decreasing = TRUE), c("term","parameter")], 4)
plot_stability_curves(g, terms = unique(top_terms$term), parameter = unique(top_terms$parameter)[1])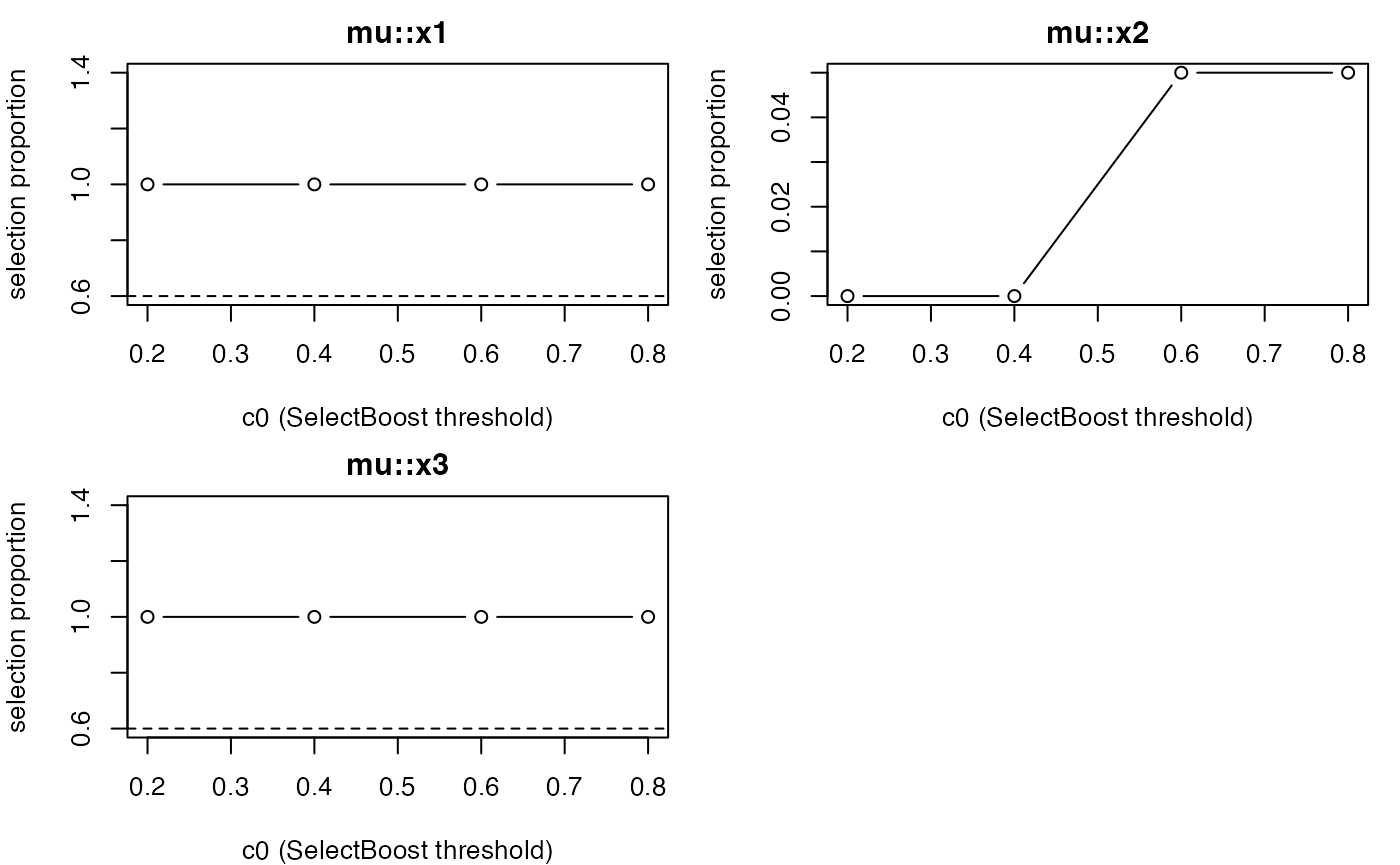
Grouped penalties for factors & splines
library(gamlss)
library(SelectBoost.gamlss)
set.seed(42)
n <- 500
f <- factor(sample(letters[1:3], n, TRUE))
x1 <- rnorm(n); x2 <- rnorm(n)
y <- gamlss.dist::rNO(n, mu = 0.3 + 1.2*x1 - 0.7*x2 + 0.5*(f=="c"), sigma = exp(0.1 + 0.2*(f=="b")))
dat <- data.frame(y, f, x1, x2)
# Stepwise baseline
fit_step <- sb_gamlss(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ f + x1 + pb(x2) + x1:f,
sigma_scope = ~ f + x1 + pb(x2),
B = 40, pi_thr = 0.6, pre_standardize = TRUE, trace = FALSE
)
# Group lasso for μ and sparse group lasso for σ
fit_grp <- sb_gamlss(
y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ f + x1 + pb(x2) + x1:f,
sigma_scope = ~ f + x1 + pb(x2),
engine = "grpreg", engine_sigma = "sgl",
B = 40, pi_thr = 0.6, pre_standardize = TRUE, trace = FALSE
)
plot_sb_gamlss(fit_step)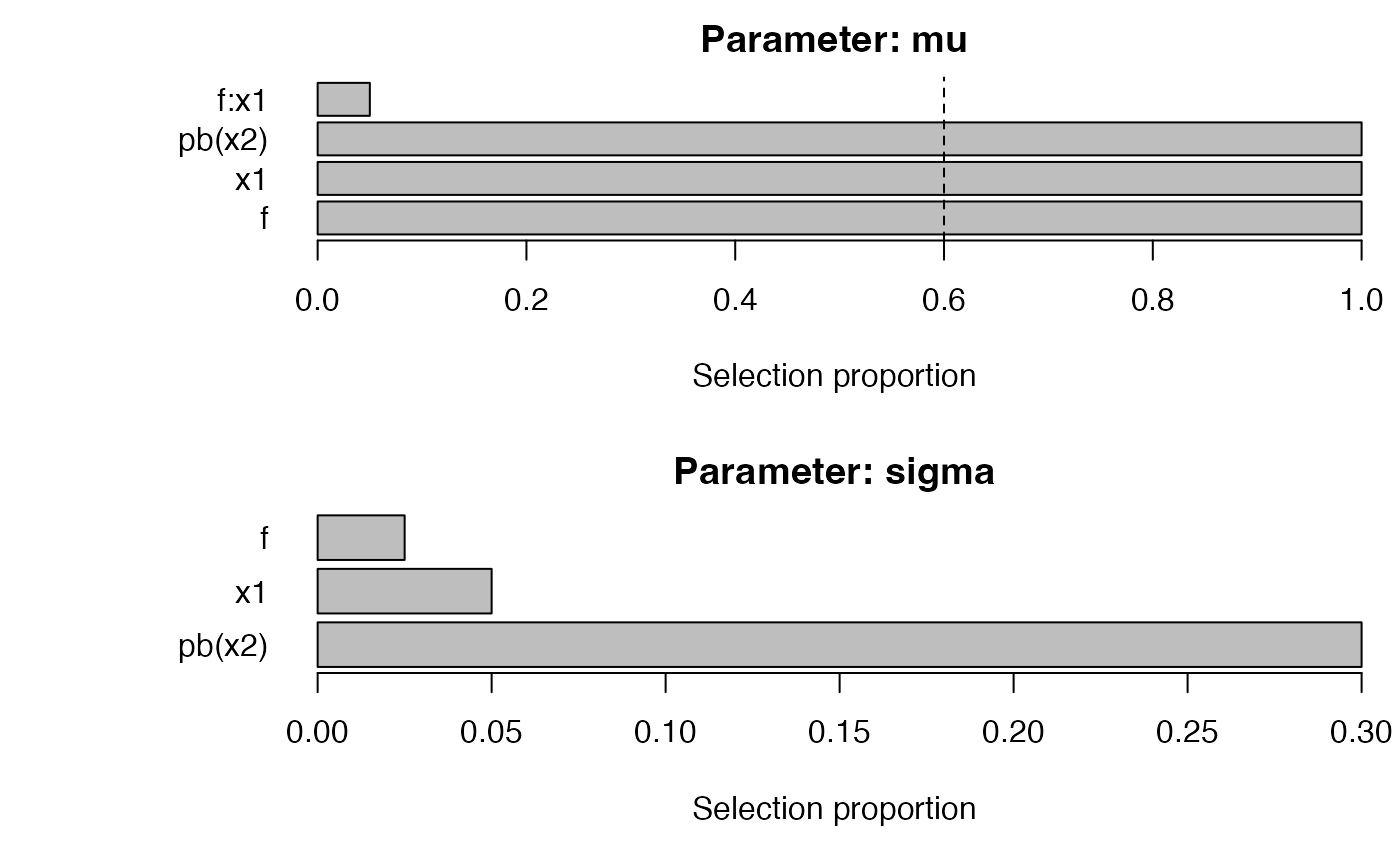
plot_sb_gamlss(fit_grp)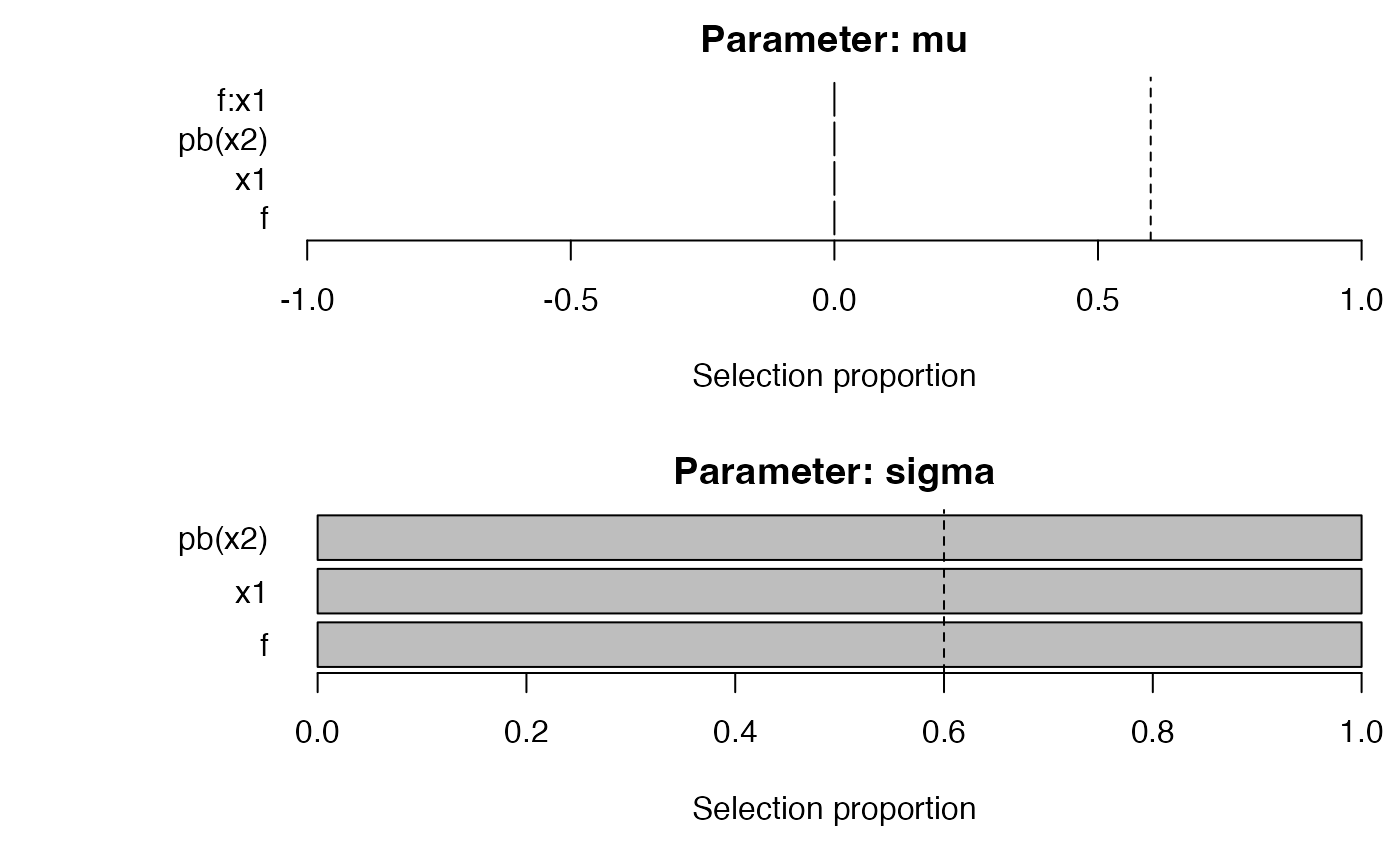
selection_table(fit_step) |> head()
#> parameter term count prop
#> 1 mu f 40 1.000
#> 2 mu x1 40 1.000
#> 3 mu pb(x2) 40 1.000
#> 4 mu f:x1 2 0.050
#> 5 sigma f 1 0.025
#> 6 sigma x1 2 0.050
selection_table(fit_grp) |> head()
#> parameter term count prop
#> 1 mu f 0 0
#> 2 mu x1 0 0
#> 3 mu pb(x2) 0 0
#> 4 mu f:x1 0 0
#> 5 sigma f 40 1
#> 6 sigma x1 40 1Tuning & Group Knockoffs
# Tuning engines / penalties
cfgs <- list(
list(engine="stepGAIC"),
list(engine="glmnet", glmnet_alpha=1),
list(engine="grpreg", grpreg_penalty="grLasso", engine_sigma="sgl", sgl_alpha=0.9)
)
base <- list(
formula = y ~ 1, data = dat, family = gamlss.dist::NO(),
mu_scope = ~ f + x1 + pb(x2) + x1:f, sigma_scope = ~ f + x1 + pb(x2),
pi_thr = 0.6, pre_standardize = TRUE, sample_fraction = 0.7, parallel = "auto", trace = FALSE
)
tuned <- tune_sb_gamlss(cfgs, base_args = base, B_small = 30)
#> | | | 0% | |======================= | 33% | |=============================================== | 67% | |======================================================================| 100%
tuned$best_config
#> $engine
#> [1] "stepGAIC"
# Approximate group knockoffs for mu
sel_mu <- NULL
if (requireNamespace("knockoff", quietly = TRUE)) {
sel_mu <- tryCatch(
knockoff_filter_mu(dat, response = "y", mu_scope = ~ f + x1 + pb(x2), fdr = 0.1),
error = function(e) {
cat("Knockoff filter failed:", conditionMessage(e), "— skipping example.\n")
NULL
}
)
} else {
cat("Package 'knockoff' not installed; skipping knockoff example.\n")
}
#> Knockoff filter failed: non-numeric argument to binary operator — skipping example.
if (!is.null(sel_mu)) sel_mu