Getting started with bigPLScox
Frédéric Bertrand
Cedric, Cnam, Parisfrederic.bertrand@lecnam.net
2025-11-16
Source:vignettes/getting-started.Rmd
getting-started.RmdWhy bigPLScox?
bigPLScox implements Partial Least Squares (PLS) extensions of the Cox proportional hazards model that remain stable in high-dimensional survival settings. The package now ships with three complementary engines:
-
coxgpls()– the classical matrix-based estimator with optional sparse and grouped variants (e.g.coxsgpls(),coxspls_sgpls()). -
big_pls_cox_fast()– the new Armadillo backend that produces variance-one components and supports both dense matrices andbigmemory::big.matrixobjects through the same interface. -
big_pls_cox_gd()– a stochastic gradient descent solver that streams data from disk and converges quickly even when only a subset of predictors remains active at each step.
Together with the deviance-residual solvers and cross-validation helpers, these functions cover the most common modelling workflows. This vignette walks through those workflows using the bundled allelotyping dataset.
Loading the example data
library(bigPLScox)
data(micro.censure)
data(Xmicro.censure_compl_imp)
Y_all <- micro.censure$survyear[1:80]
status_all <- micro.censure$DC[1:80]
X_all <- apply(
as.matrix(Xmicro.censure_compl_imp),
MARGIN = 2,
FUN = as.numeric
)[1:80, ]
set.seed(2024)
train_id <- 1:60
test_id <- 61:80
X_train <- X_all[train_id, ]
X_test <- X_all[test_id, ]
Y_train <- Y_all[train_id]
Y_test <- Y_all[test_id]
status_train <- status_all[train_id]
status_test <- status_all[test_id]The original factor-based design matrix is also available if you wish to leverage the formula interface.
X_train_raw <- Xmicro.censure_compl_imp[train_id, ]
X_test_raw <- Xmicro.censure_compl_imp[test_id, ]Inspecting deviance residuals
Deviance residuals can reveal problematic observations before any
components are extracted. The helper computeDR() exposes
both an R and a C++ engine; the latter is substantially faster and
powers the new fast PLS routines.
residuals_overview <- computeDR(Y_train, status_train, plot = TRUE)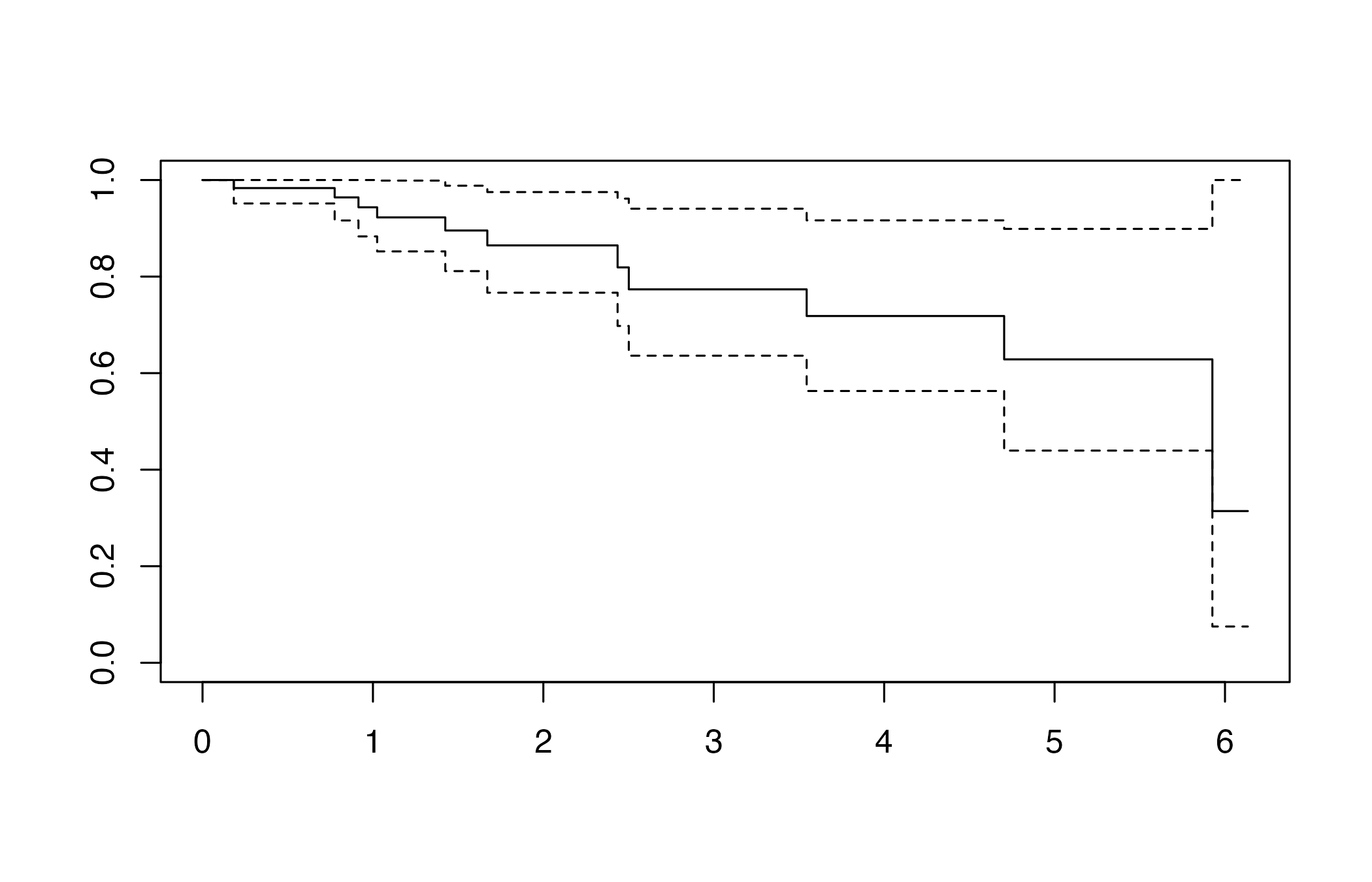
eta_null <- rep(0, length(Y_train))
head(residuals_overview)
#> 1 2 3 4 5 6
#> -1.3771591 -0.5360370 -0.2693493 -0.3994329 -0.8040940 -0.3994329
if (requireNamespace("bench", quietly = TRUE)) {
benchmark_dr <- bench::mark(
survival = computeDR(Y_train, status_train, engine = "survival"),
cpp = computeDR(Y_train, status_train, engine = "cpp", eta = eta_null),
iterations = 10,
check = FALSE
)
benchmark_dr
}
#> # A tibble: 2 × 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 survival 813.7µs 855.5µs 1136. 169KB 0
#> 2 cpp 88.7µs 91.9µs 10259. 163KB 0
all.equal(
as.numeric(computeDR(Y_train, status_train, engine = "survival")),
as.numeric(computeDR(Y_train, status_train, engine = "cpp", eta = eta_null)),
tolerance = 1e-7
)
#> [1] TRUEMatrix-based PLS–Cox models
The matrix interface mimics survival::coxph() while
augmenting the predictor space with latent components. Most users start
with coxgpls() and the cross-validation helper
cv.coxgpls().
set.seed(123)
cox_pls_fit <- coxgpls(
Xplan = X_train,
time = Y_train,
status = status_train,
ncomp = 6,
ind.block.x = c(3, 10, 20)
)
cox_pls_fit
#> Call:
#> coxph(formula = YCsurv ~ ., data = tt_gpls)
#>
#> coef exp(coef) se(coef) z p
#> dim.1 -0.7368 0.4786 0.1162 -6.340 2.3e-10
#> dim.2 -0.5256 0.5912 0.1382 -3.804 0.000142
#> dim.3 -0.3314 0.7179 0.1199 -2.763 0.005720
#> dim.4 -0.2883 0.7495 0.1092 -2.641 0.008272
#> dim.5 -0.4002 0.6702 0.1435 -2.788 0.005298
#> dim.6 -0.2696 0.7636 0.1239 -2.176 0.029529
#>
#> Likelihood ratio test=60.94 on 6 df, p=2.906e-11
#> n= 60, number of events= 60The formula interface accepts data frames containing the predictors along with survival outcomes.
cox_pls_fit_formula <- coxgpls(
~ ., Y_train, status_train,
ncomp = 6,
ind.block.x = c(3, 10, 20),
dataXplan = data.frame(X_train_raw)
)
cox_pls_fit_formula
#> Call:
#> coxph(formula = YCsurv ~ ., data = tt_gpls)
#>
#> coef exp(coef) se(coef) z p
#> dim.1 -0.82612 0.43775 0.31903 -2.589 0.00961
#> dim.2 -0.79075 0.45350 0.39461 -2.004 0.04508
#> dim.3 -0.89888 0.40703 0.30294 -2.967 0.00301
#> dim.4 0.02354 1.02382 0.29663 0.079 0.93675
#> dim.5 -0.40714 0.66555 0.40456 -1.006 0.31423
#> dim.6 -0.53689 0.58456 0.38554 -1.393 0.16374
#>
#> Likelihood ratio test=19.59 on 6 df, p=0.00328
#> n= 60, number of events= 11Cross-validation
Repeated cross-validation stabilises the choice of latent components. The returned object records the optimal number of components and diagnostic curves.
set.seed(123456)
cv_results <- suppressWarnings(cv.coxgpls(
list(x = X_train, time = Y_train, status = status_train),
nt = 6,
ind.block.x = c(3, 10, 20)
))
#> CV Fold 1
#> CV Fold 2
#> CV Fold 3
#> CV Fold 4
#> CV Fold 5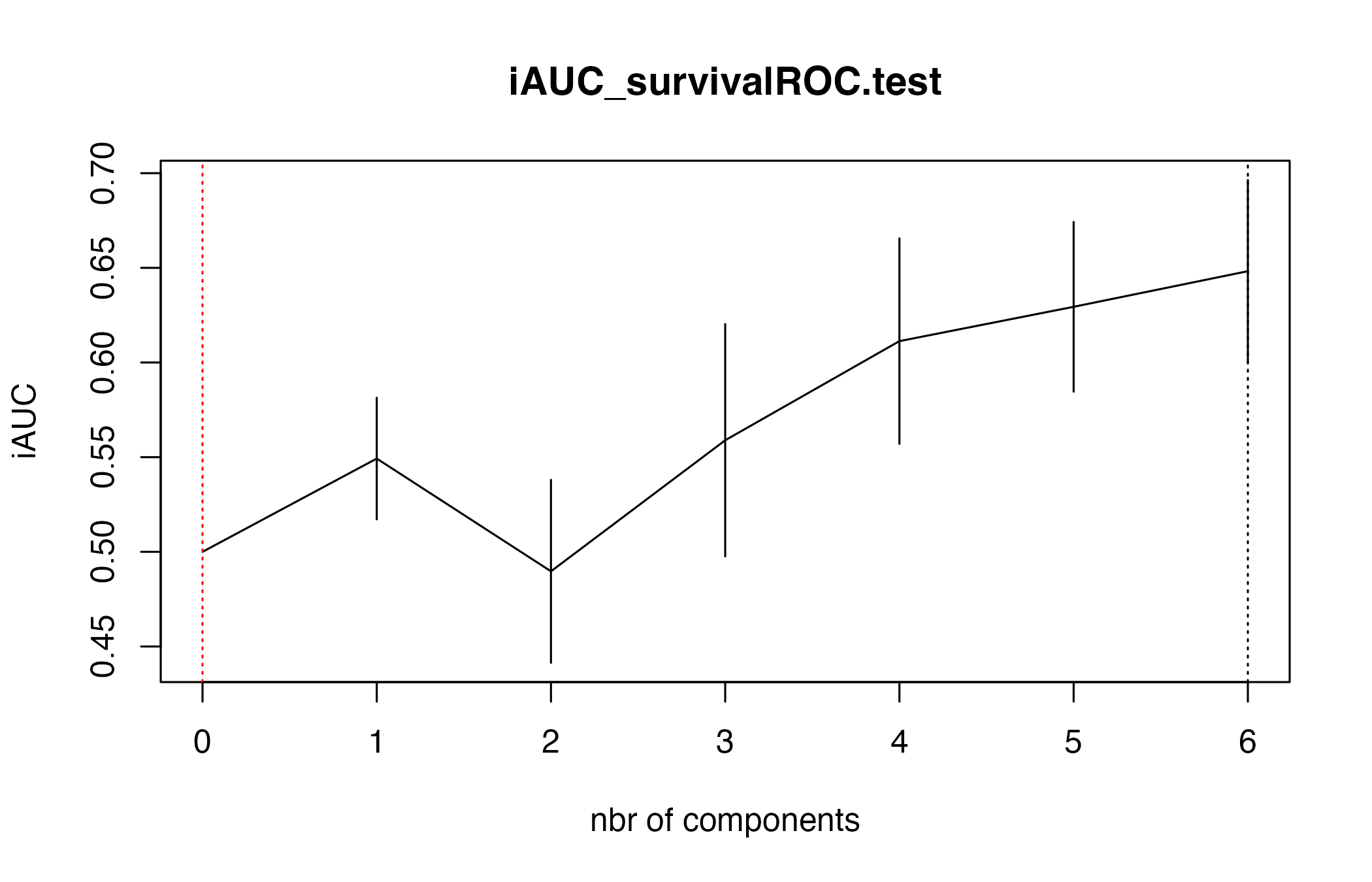
cv_results
#> $nt
#> [1] 6
#>
#> $cv.error10
#> [1] 0.5000000 0.5492633 0.4897065 0.5589258 0.6112917 0.6294183 0.6482323
#>
#> $cv.se10
#> [1] 0.00000000 0.03211886 0.04830433 0.06137605 0.05429528 0.04481718 0.04814978
#>
#> $folds
#> $folds$`1`
#> [1] 60 45 3 57 21 15 35 22 51 12 20 13
#>
#> $folds$`2`
#> [1] 42 54 50 28 1 41 6 18 44 8 27 25
#>
#> $folds$`3`
#> [1] 59 36 55 52 24 46 37 19 4 47 33 5
#>
#> $folds$`4`
#> [1] 49 38 30 2 34 48 53 31 11 56 26 39
#>
#> $folds$`5`
#> [1] 7 10 23 16 14 58 29 9 43 17 40 32
#>
#>
#> $lambda.min10
#> [1] 6
#>
#> $lambda.1se10
#> [1] 0Use the selected number of components to refit with the deviance-residual solver for a robustness check.
set.seed(123456)
cox_pls_dr <- coxgplsDR(
Xplan = X_train,
time = Y_train,
status = status_train,
ncomp = cv_results$nt,
ind.block.x = c(3, 10, 20)
)
cox_pls_dr
#> Call:
#> coxph(formula = YCsurv ~ ., data = tt_gplsDR)
#>
#> coef exp(coef) se(coef) z p
#> dim.1 0.7329 2.0812 0.1120 6.545 5.95e-11
#> dim.2 0.6418 1.8999 0.1456 4.409 1.04e-05
#> dim.3 0.3467 1.4144 0.1080 3.210 0.00133
#> dim.4 0.4266 1.5320 0.1554 2.745 0.00605
#> dim.5 0.3694 1.4468 0.1453 2.542 0.01101
#> dim.6 0.2884 1.3343 0.1095 2.633 0.00847
#>
#> Likelihood ratio test=63.84 on 6 df, p=7.442e-12
#> n= 60, number of events= 60Sparse and structured sparse variants (coxsgpls(),
coxspls_sgpls()) share the same workflow with additional
arguments that control the number of selected predictors per component
(keepX) or penalty strength.
Fast solvers for medium-sized data
The new big_pls_cox_fast() routine exposes identical
arguments for dense matrices and big.matrix objects. On
moderate data it serves as a drop-in replacement for the original R
implementation big_pls_cox().
fast_fit_dense <- big_pls_cox_fast(
X = X_train,
time = Y_train,
status = status_train,
ncomp = 4
)
summary(fast_fit_dense)
#> $n
#> [1] 60
#>
#> $p
#> [1] 40
#>
#> $ncomp
#> [1] 4
#>
#> $keepX
#> [1] 0 0 0 0
#>
#> $center
#> [1] 0.4666667 0.4500000 0.4166667 0.5333333 0.5000000 0.5166667
#> [7] 0.4833333 0.4000000 0.3833333 0.4500000 0.5166667 0.5166667
#> [13] 0.5500000 0.3666667 0.4333333 0.5333333 0.4833333 0.5166667
#> [19] 0.5500000 0.5166667 0.4333333 0.5666667 0.4000000 0.3833333
#> [25] 0.5166667 0.5166667 0.5666667 0.5333333 0.4666667 0.4500000
#> [31] 0.5500000 0.5500000 0.4500000 0.6333333 62.4622365 1.6833333
#> [37] 2.3833333 0.5166667 0.2500000 2.2666667
#>
#> $scale
#> [1] 0.5030977 0.5016921 0.4971671 0.5030977 0.5042195 0.5039393
#> [7] 0.5039393 0.4940322 0.4903014 0.5016921 0.5039393 0.5039393
#> [13] 0.5016921 0.4859611 0.4997174 0.5030977 0.5039393 0.5039393
#> [19] 0.5016921 0.5039393 0.4997174 0.4997174 0.4940322 0.4903014
#> [25] 0.5039393 0.5039393 0.4997174 0.5030977 0.5030977 0.5016921
#> [31] 0.5016921 0.5016921 0.5016921 0.4859611 14.6024282 0.7008873
#> [37] 0.9404591 0.7917299 0.4366669 1.2604367
#>
#> $cox
#> Call:
#> survival::coxph(formula = survival::Surv(time, status) ~ ., data = scores_df,
#> ties = "efron", x = FALSE)
#>
#> n= 60, number of events= 11
#>
#> coef exp(coef) se(coef) z Pr(>|z|)
#> comp1 -1.4836 0.2268 0.5002 -2.966 0.00301 **
#> comp2 1.3937 4.0299 0.5081 2.743 0.00609 **
#> comp3 -1.4271 0.2400 0.6997 -2.040 0.04138 *
#> comp4 0.9357 2.5490 0.5119 1.828 0.06755 .
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> exp(coef) exp(-coef) lower .95 upper .95
#> comp1 0.2268 4.4088 0.0851 0.6045
#> comp2 4.0299 0.2481 1.4887 10.9090
#> comp3 0.2400 4.1668 0.0609 0.9458
#> comp4 2.5490 0.3923 0.9347 6.9518
#>
#> Concordance= 0.962 (se = 0.015 )
#> Likelihood ratio test= 40.45 on 4 df, p=3e-08
#> Wald test = 15.58 on 4 df, p=0.004
#> Score (logrank) test = 38.25 on 4 df, p=1e-07
#>
#>
#> attr(,"class")
#> [1] "summary.big_pls_cox"Predictions rely on the same predict() interface used by
the classical function.
linear_predictor_fast <- predict(fast_fit_dense, newdata = X_test, type = "link")
head(linear_predictor_fast)
#> [1] 3.3499156 -0.8685314 8.6594617 -6.5718861 2.0102711 -2.1990051For comparison, the legacy solver is still available. The C++ backend usually reduces runtime by an order of magnitude while delivering components scaled to variance one.
legacy_fit_dense <- big_pls_cox(
X = X_train,
time = Y_train,
status = status_train,
ncomp = 4
)
legacy_fit_dense$cox_fit
#> $coefficients
#> [1] 6.200427 3.171981 -2.748535 1.293296
#>
#> $var
#> [,1] [,2] [,3] [,4]
#> [1,] 5.821149 2.4185955 -2.3040552 1.0187707
#> [2,] 2.418596 1.6445800 -1.1764804 0.5610664
#> [3,] -2.304055 -1.1764804 1.2404846 -0.5308908
#> [4,] 1.018771 0.5610664 -0.5308908 0.3210964
#>
#> $loglik
#> [1] -33.823093 -6.374114
#>
#> $score
#> [1] 34.35164
#>
#> $iter
#> [1] 8
#>
#> $linear.predictors
#> [1] -12.57620642 -0.80958795 3.92609801 2.60017390 1.76853806
#> [6] -3.36295838 -1.07944356 10.31904141 -3.64756309 -2.89617173
#> [11] 10.99977612 -3.67120048 0.19277160 -4.93349776 0.72346177
#> [16] -2.96898338 -11.92153226 -16.36665206 7.29677779 -6.25230861
#> [21] -7.44252580 -3.35748178 0.89833874 -0.03190649 -14.18767386
#> [26] -5.70848324 10.05566087 2.33425751 1.44427382 -9.42823092
#> [31] 16.81484345 -8.16109725 -1.01887152 1.67809982 9.52923393
#> [36] -0.81577205 13.21271969 4.03431692 -8.22578371 8.37431879
#> [41] 8.42795290 2.19412589 -1.95037872 2.94403579 -3.81221056
#> [46] -0.29923400 -13.50366653 1.71522979 6.77552365 -1.04786702
#> [51] -1.05373010 3.52071260 4.14906575 13.53253117 16.06054363
#> [56] -9.18209817 -14.09511037 6.91845019 0.66507554 0.70227868
#>
#> $residuals
#> 1 2 3 4 5
#> -9.951099e-06 -1.028552e-05 -3.995987e-06 -2.023840e-05 -5.242897e-03
#> 6 7 8 9 10
#> -5.204999e-08 -5.106679e-07 2.996282e-01 -6.021565e-07 -1.784430e-09
#> 11 12 13 14 15
#> 9.535666e-01 -2.005207e-09 -1.822446e-06 -2.171991e-04 -6.660185e-08
#> 16 17 18 19 20
#> -1.659120e-09 -1.535926e-10 -1.172164e-13 -3.196396e-01 -5.809057e-05
#> 21 22 23 24 25
#> -1.824357e-09 -2.701177e-08 -3.690443e-06 -8.662599e-04 -2.078836e-08
#> 26 27 28 29 30
#> -1.071835e-10 -7.252299e-02 -3.214496e-05 -6.370523e-06 -6.337423e-12
#> 31 32 33 34 35
#> -5.817400e-01 -6.599430e-09 -8.343257e-06 -1.307490e-03 -1.851665e-02
#> 36 37 38 39 40
#> -3.955667e-04 -7.043898e-01 -1.305893e-03 -8.647257e-12 -5.820366e-02
#> 41 42 43 44 45
#> -1.477421e-04 -2.073589e-04 -2.137451e-07 -1.698570e-02 -7.139517e-10
#> 46 47 48 49 50
#> -1.810061e-04 -4.119846e-08 -1.730902e-05 -1.316762e-03 -1.056724e-02
#> 51 52 53 54 55
#> -8.511766e-05 -1.970571e-02 -9.524699e-05 -1.325008e-01 6.950025e-01
#> 56 57 58 59 60
#> -2.377340e-09 -2.280448e-08 -3.265394e-05 -4.494326e-05 -1.805098e-03
#>
#> $means
#> [1] 3.330669e-17 -4.070818e-17 -4.996004e-17 3.700743e-18
#>
#> $method
#> [1] "efron"
#>
#> $class
#> [1] "coxph"Fast PLS Cox vs gradient based PLS Cox
The package provides two families of PLS Cox estimators:
-
big_pls_cox_fast
Exact PLS components computed in compiled code, with a single Cox fit at the end. -
big_pls_cox_gd
A gradient based Cox optimisation performed in the latent PLS space.
Both approaches share the same preprocessing (centering and scaling), the same PLS deflation scheme and the same prediction interface. The main difference is how the final Cox coefficients are obtained.
In the exact fast path, we build PLS components that are tailored to Cox score residuals, then fit a Cox model once on the resulting scores. In the gradient based path, we optimise the partial log-likelihood directly in the space spanned by the PLS scores, using one of several optimisers.
The code below illustrates a typical benchmark on a moderate survival data set. The design is split into a training set used to fit the models and a test set used to evaluate concordance.
set.seed(2024)
# Exact fast PLS Cox (dense)
fast_fit_dense <- big_pls_cox_fast(
X = X_train,
time = Y_train,
status = status_train,
ncomp = 4
)
# Exact fast PLS Cox (big.matrix)
if (requireNamespace("bigmemory", quietly = TRUE)) {
library(bigmemory)
X_big_train <- bigmemory::as.big.matrix(X_train)
X_big_test <- bigmemory::as.big.matrix(X_test)
fast_fit_big <- big_pls_cox_fast(
X = X_big_train,
time = Y_train,
status = status_train,
ncomp = 4
)
# Gradient based fit in the same latent space
gd_fit <- big_pls_cox_gd(
X = X_big_train,
time = Y_train,
status = status_train,
ncomp = 4,
max_iter = 2000
)
risk_table <- data.frame(
subject = seq_along(test_id),
fast_dense = predict(fast_fit_dense, newdata = X_test, type = "link"),
fast_big = predict(fast_fit_big, newdata = X_big_test, type = "link"),
gd = predict(gd_fit, newdata = X_big_test, type = "link")
)
concordances <- apply(
risk_table[-1],
2,
function(lp) {
survival::concordance(
survival::Surv(Y_test, status_test) ~ lp
)$concordance
}
)
concordances
}
#> fast_dense fast_big gd
#> 0.4418605 0.4418605 0.2790698big_pls_cox_gd() is particularly useful for streamed
data or when the number of active predictors per component is restricted
via keepX.
Predictions and evaluation
All solvers return latent scores and loadings that can be reused for
plotting or external validation. Use
predict(..., type = "components") to extract the scores
directly.
if (exists("fast_fit_dense")) {
component_scores <- predict(fast_fit_dense, newdata = X_test, type = "components")
head(component_scores)
}
#> [,1] [,2] [,3] [,4]
#> [1,] -0.61328247 1.45903382 -0.2024545 0.1256753
#> [2,] -0.14585655 -0.89077212 -0.3630109 -0.3863126
#> [3,] -2.49681489 1.07906464 -1.7437503 1.0288135
#> [4,] -0.01412341 -2.48245825 1.7023219 -0.7517977
#> [5,] -0.56984205 -0.10012954 -1.0183793 -0.1591913
#> [6,] 0.49642902 -0.09180402 0.5767821 -0.5465379The concordance index provides a quick check of predictive ability on the test set.
if (exists("fast_fit_dense")) {
concordance_fast <- survival::concordance(
survival::Surv(Y_test, status_test) ~ linear_predictor_fast
)$concordance
concordance_fast
}
#> [1] 0.4418605DK-splines extension
For flexible baseline hazards the coxDKgplsDR()
estimator augments the PLS components with DK-splines. The interface
mirrors the previous functions.
cox_DKsplsDR_fit <- coxDKgplsDR(
Xplan = X_train,
time = Y_train,
status = status_train,
ncomp = 6,
validation = "CV",
ind.block.x = c(3, 10, 20),
verbose = FALSE
)
cox_DKsplsDR_fit
#> Call:
#> coxph(formula = YCsurv ~ ., data = tt_DKgplsDR)
#>
#> coef exp(coef) se(coef) z p
#> dim.1 4.4294 83.8841 0.7310 6.060 1.36e-09
#> dim.2 3.6711 39.2955 0.8183 4.487 7.24e-06
#> dim.3 3.8127 45.2712 1.0314 3.697 0.000218
#> dim.4 2.6709 14.4534 0.8963 2.980 0.002883
#> dim.5 1.6486 5.1998 0.6897 2.390 0.016827
#> dim.6 1.4384 4.2139 0.7022 2.048 0.040517
#>
#> Likelihood ratio test=66.07 on 6 df, p=2.612e-12
#> n= 60, number of events= 60Cross-validation is available for the DK-splines estimator as well.
set.seed(2468)
cv_coxDKgplsDR_res <- suppressWarnings(cv.coxDKgplsDR(
list(x = X_train, time = Y_train, status = status_train),
nt = 6,
ind.block.x = c(3, 10, 20)
))
#> Kernel : rbfdot
#> Estimated_sigma 0.01258323
#> CV Fold 1
#> Kernel : rbfdot
#> Estimated_sigma 0.01471071
#> CV Fold 2
#> Kernel : rbfdot
#> Estimated_sigma 0.0127949
#> CV Fold 3
#> Kernel : rbfdot
#> Estimated_sigma 0.0122611
#> CV Fold 4
#> Kernel : rbfdot
#> Estimated_sigma 0.01289496
#> CV Fold 5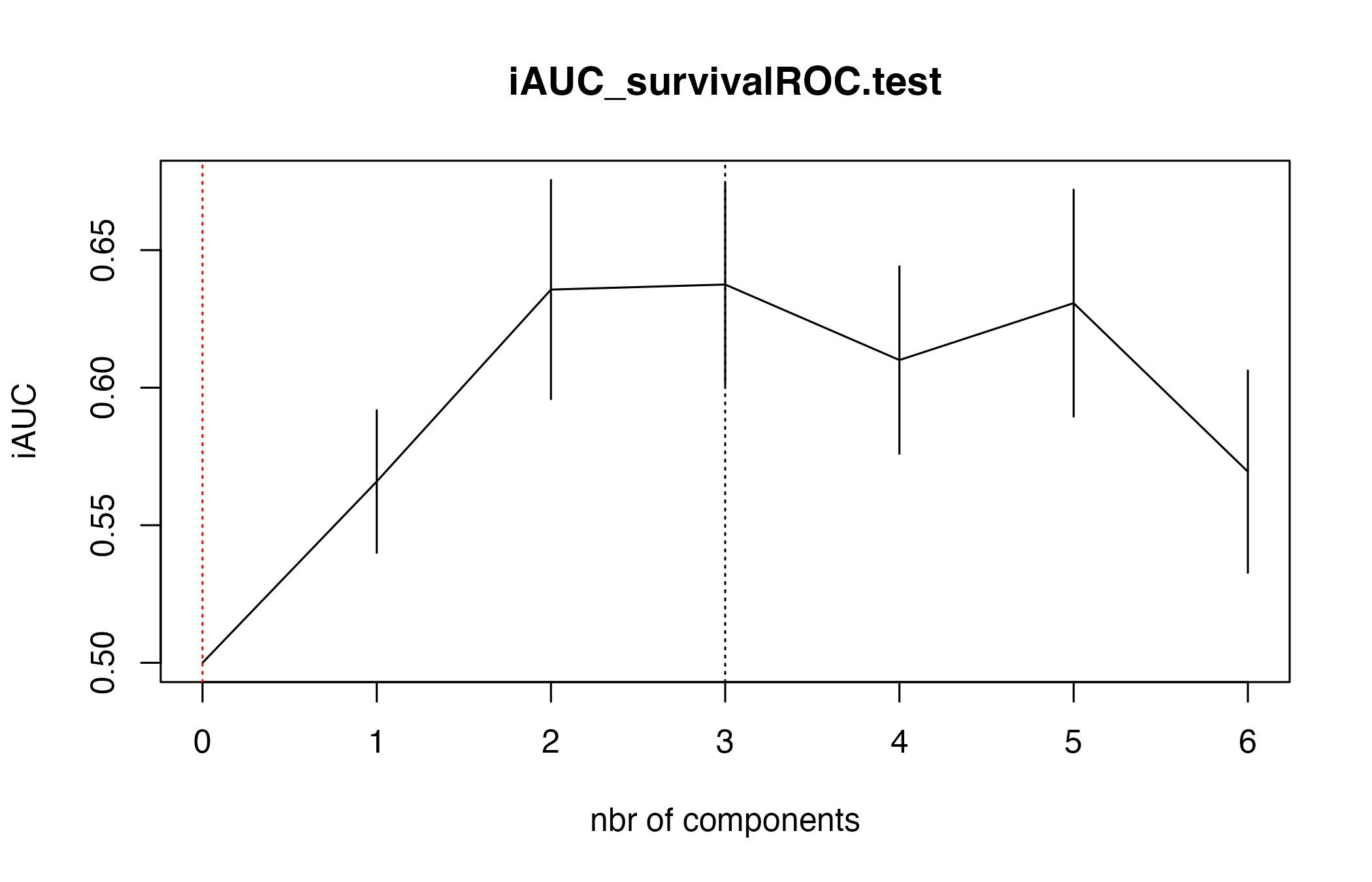
cv_coxDKgplsDR_res
#> $nt
#> [1] 6
#>
#> $cv.error10
#> [1] 0.5000000 0.5658906 0.6356608 0.6374963 0.6100371 0.6307320 0.5694802
#>
#> $cv.se10
#> [1] 0.00000000 0.02591444 0.03982352 0.03646931 0.03407857 0.04124367 0.03676530
#>
#> $folds
#> $folds$`1`
#> [1] 44 16 27 26 55 20 49 14 6 47 18 7
#>
#> $folds$`2`
#> [1] 58 8 2 17 3 15 52 43 56 11 29 59
#>
#> $folds$`3`
#> [1] 51 13 57 46 45 32 19 5 36 33 10 28
#>
#> $folds$`4`
#> [1] 21 50 38 60 53 42 23 31 12 24 1 25
#>
#> $folds$`5`
#> [1] 22 39 35 54 4 30 34 48 37 40 9 41
#>
#>
#> $lambda.min10
#> [1] 3
#>
#> $lambda.1se10
#> [1] 0Next steps
-
vignette("bigPLScox", package = "bigPLScox")dives deeper into the fast big-memory backends and shows how to reconcile the dense and streaming implementations. -
vignette("bigPLScox-benchmarking", package = "bigPLScox")demonstrates a reproducible benchmarking workflow that contrasts the classical, fast, and gradient-descent solvers againstsurvival::coxph(). - The reference documentation (
help("big_pls_cox_fast"), etc.) details every argument and return value for the modelling functions discussed above.