This function plots the confidence intervals derived using the function
confints.bootpls from from a bootpls based object.
Arguments
- ic_bootobject
an object created with the
confints.bootplsfunction.- indices
vector of indices of the variables to plot. Defaults to
NULL: all the predictors will be used.- legendpos
position of the legend as in
legend, defaults to"topleft"- prednames
do the original names of the predictors shall be plotted ? Defaults to
TRUE: the names are plotted.- articlestyle
do the extra blank zones of the margin shall be removed from the plot ? Defaults to
TRUE: the margins are removed.- xaxisticks
do ticks for the x axis shall be plotted ? Defaults to
TRUE: the ticks are plotted.- ltyIC
lty as in
plot- colIC
col as in
plot- typeIC
type of CI to plot. Defaults to
typeIC=c("Normal", "Basic", "Percentile", "BCa")if BCa intervals limits were computed and totypeIC=c("Normal", "Basic", "Percentile")otherwise.- las
numeric in 0,1,2,3; the style of axis labels. 0: always parallel to the axis [default], 1: always horizontal, 2: always perpendicular to the axis, 3: always vertical.
- mar
A numerical vector of the form
c(bottom, left, top, right)which gives the number of lines of margin to be specified on the four sides of the plot. The default isc(5, 4, 4, 2) + 0.1.- mgp
The margin line (in mex units) for the axis title, axis labels and axis line. Note that
mgp[1]affects title whereasmgp[2:3]affect axis. The default isc(3, 1, 0).- ...
further options to pass to the
plotfunction.
Author
Frédéric Bertrand
frederic.bertrand@lecnam.net
https://fbertran.github.io/homepage/
Examples
data(Cornell)
modpls <- plsR(Y~.,data=Cornell,3)
#> ____************************************************____
#> ____Component____ 1 ____
#> ____Component____ 2 ____
#> ____Component____ 3 ____
#> ____Predicting X without NA neither in X nor in Y____
#> ****________________________________________________****
#>
# Lazraq-Cleroux PLS (Y,X) bootstrap
set.seed(250)
Cornell.bootYX <- bootpls(modpls, R=250, verbose=FALSE)
temp.ci <- confints.bootpls(Cornell.bootYX,2:8)
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
plots.confints.bootpls(temp.ci)
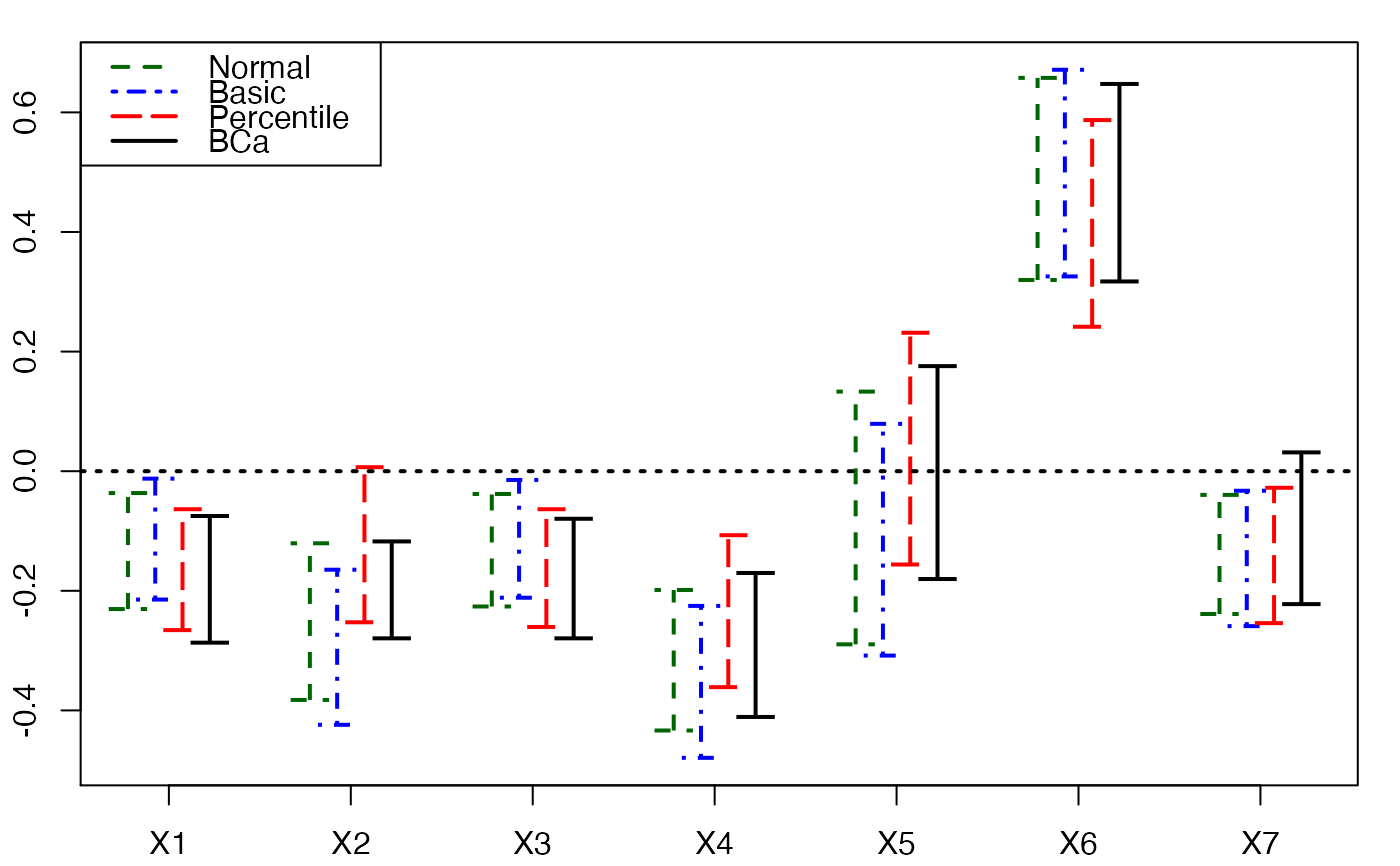 plots.confints.bootpls(temp.ci,prednames=FALSE)
plots.confints.bootpls(temp.ci,prednames=FALSE)
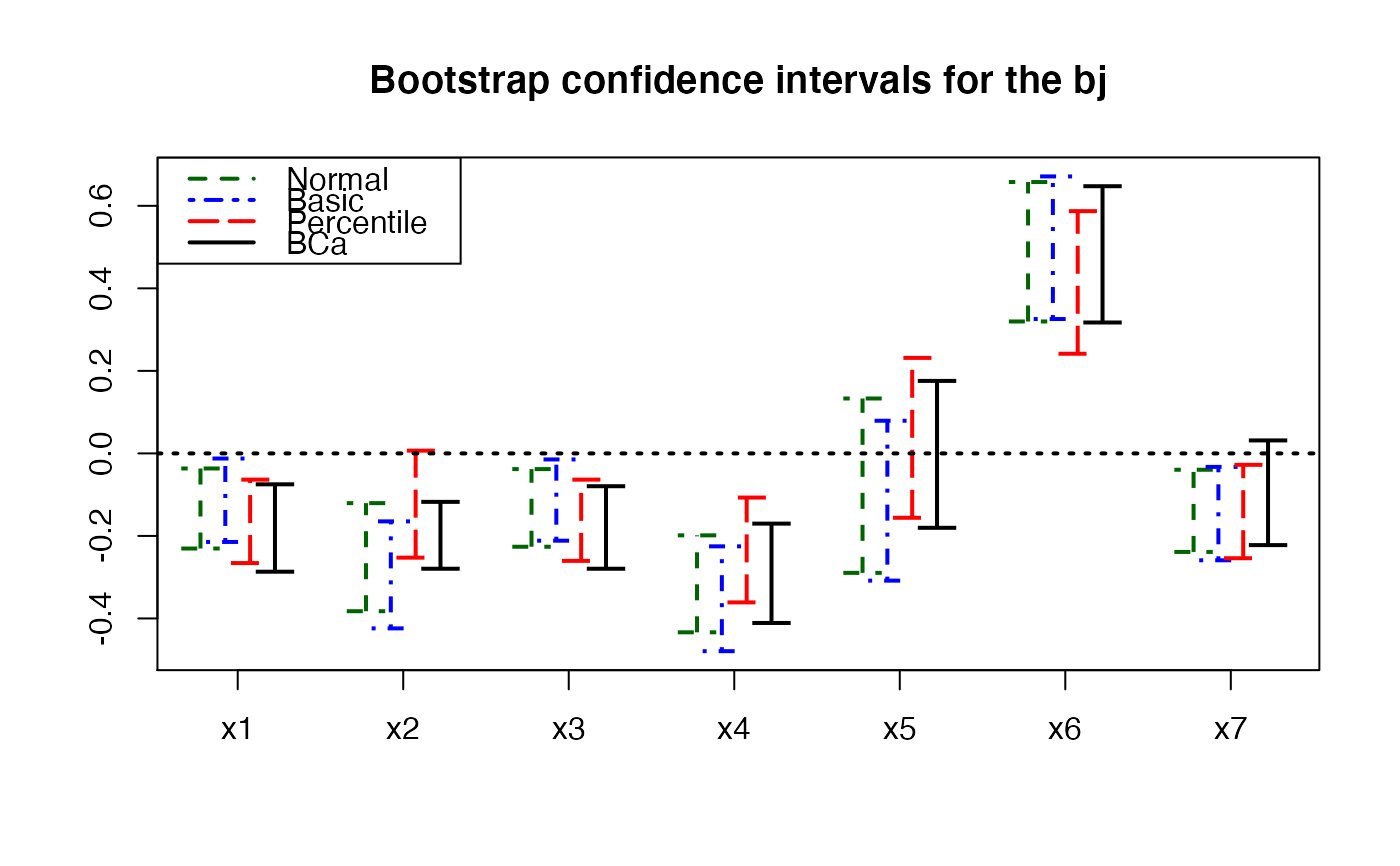
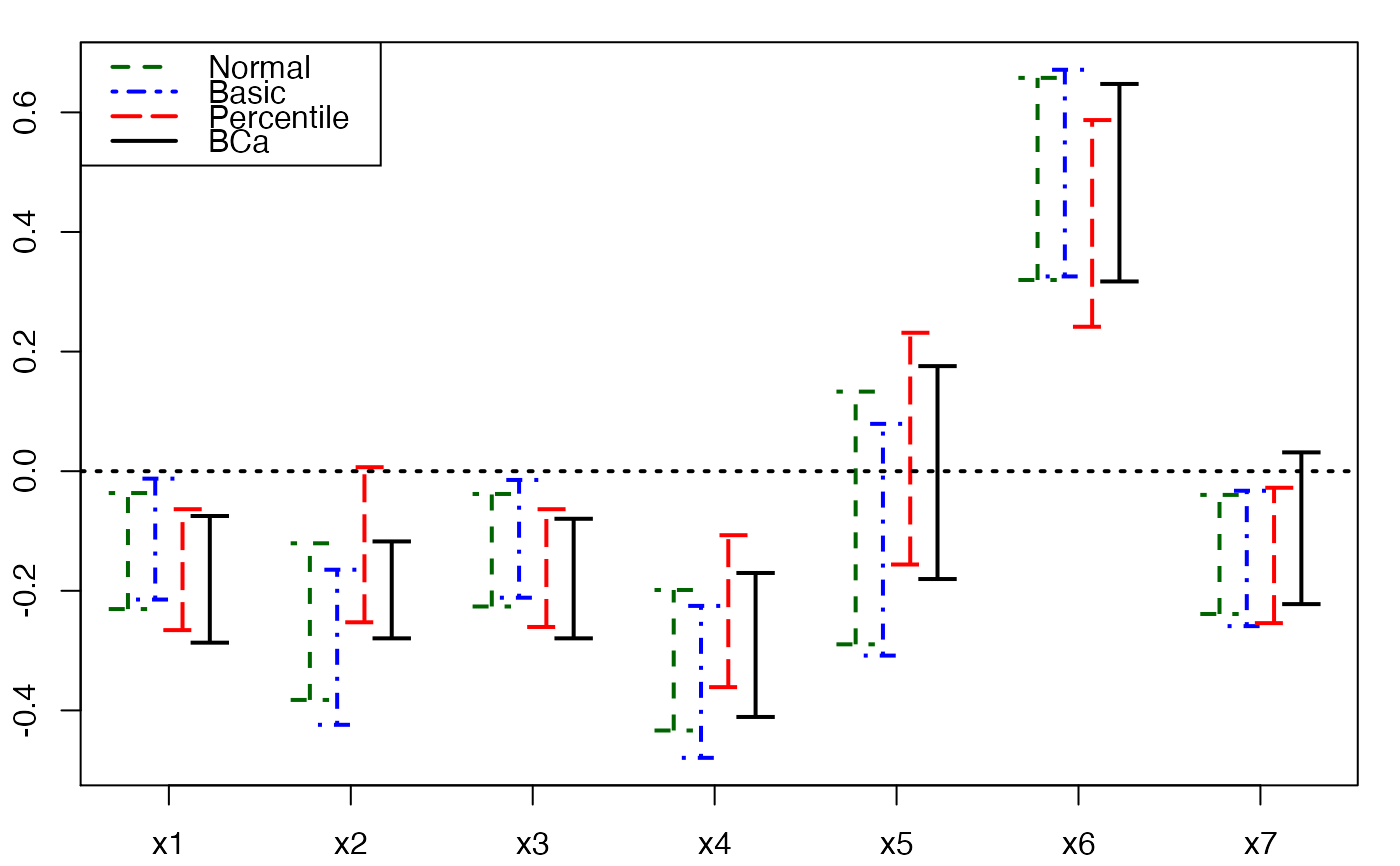 plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
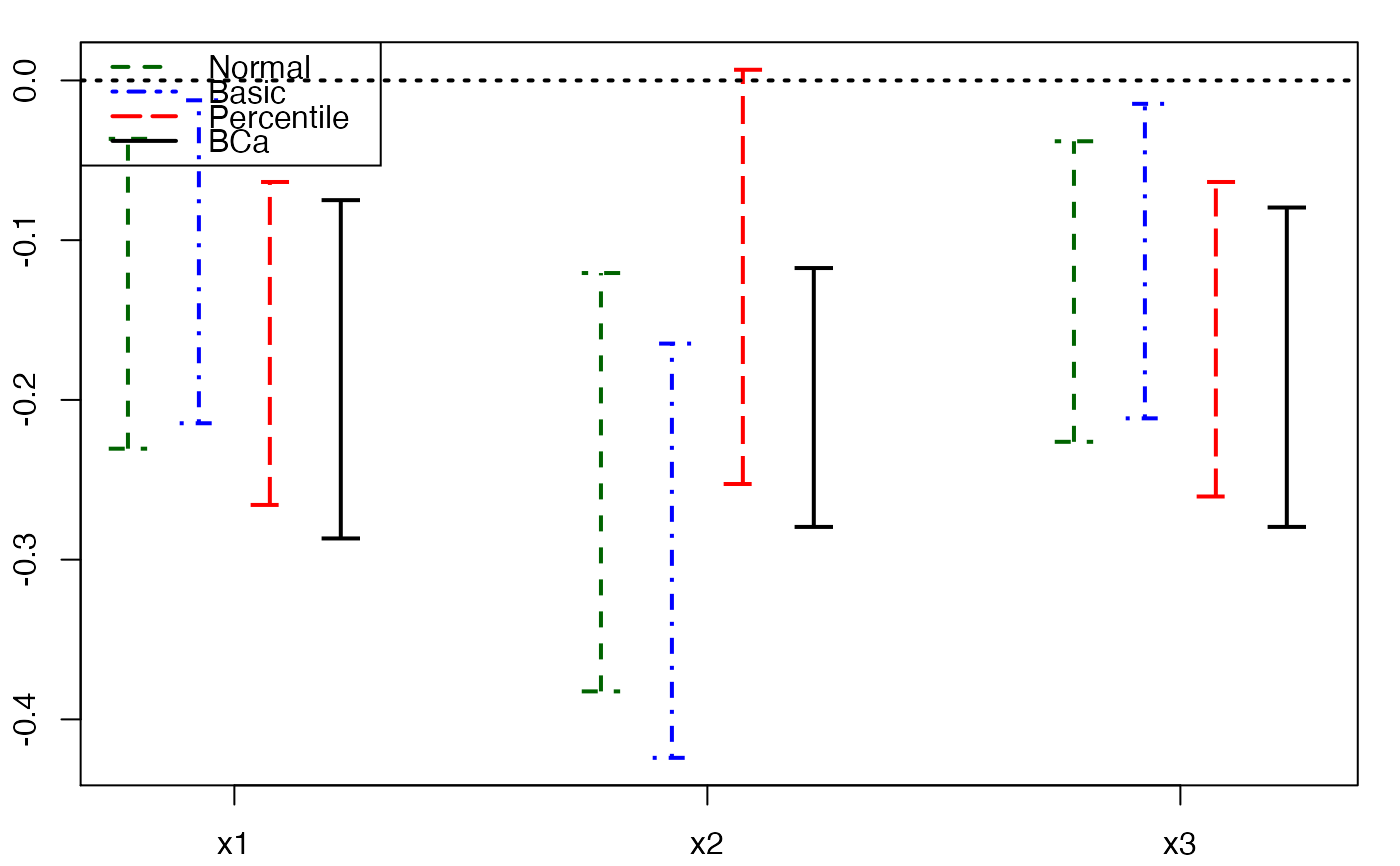
 plots.confints.bootpls(temp.ci,indices=1:3,prednames=FALSE)
plots.confints.bootpls(temp.ci,indices=1:3,prednames=FALSE)
 plots.confints.bootpls(temp.ci,c(2,4,6),"bottomright")
plots.confints.bootpls(temp.ci,c(2,4,6),"bottomright")
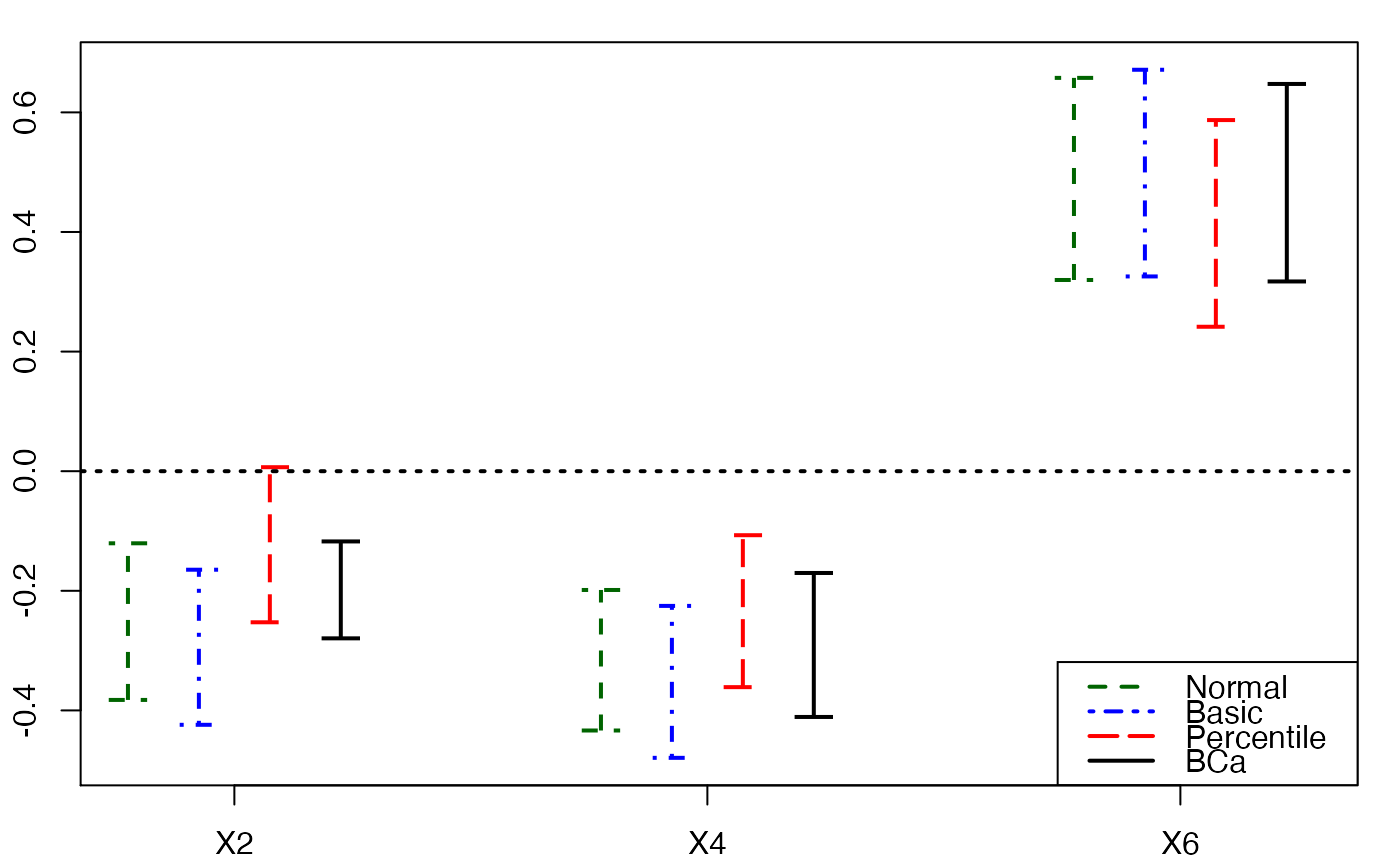 plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
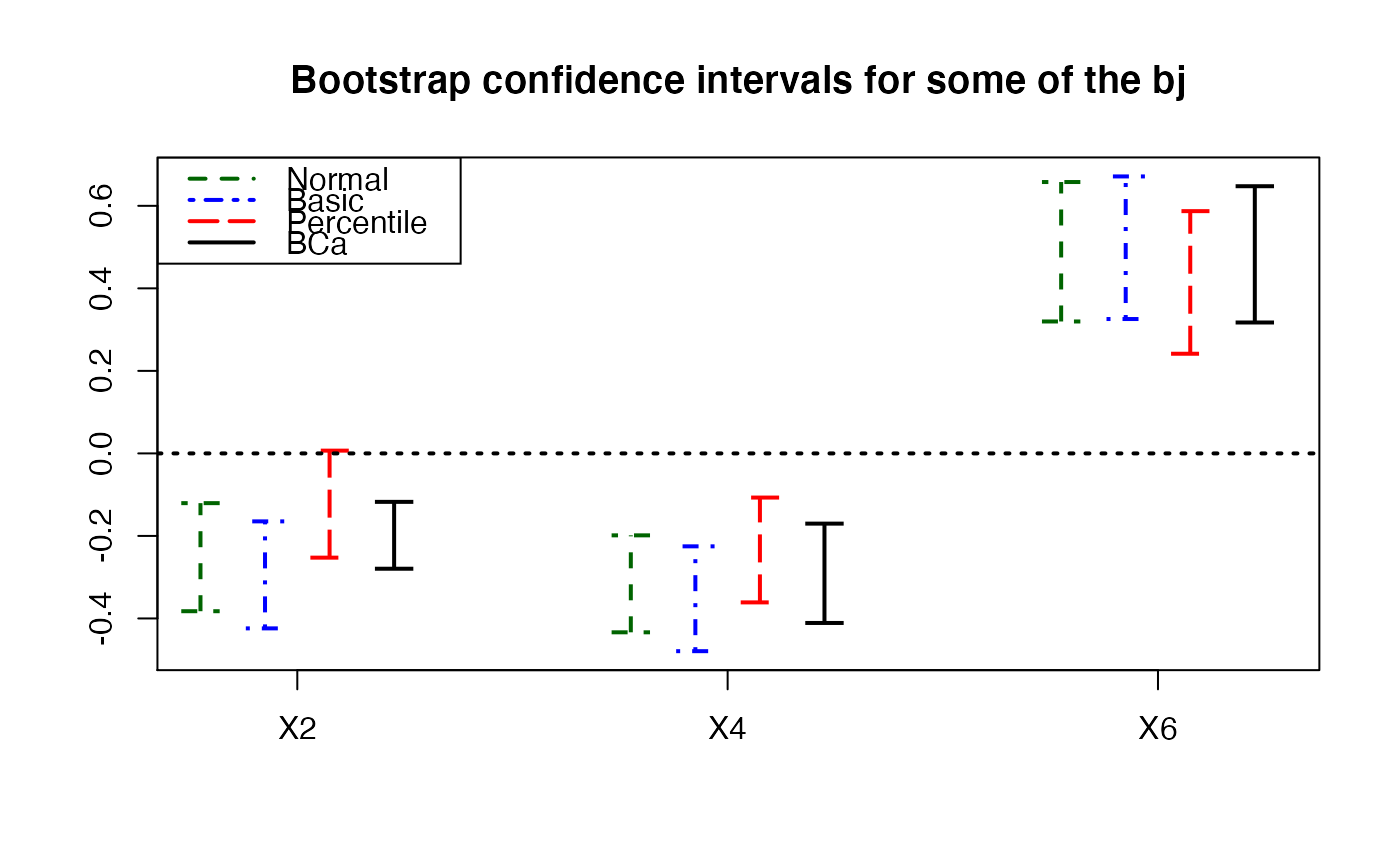 temp.ci <- confints.bootpls(Cornell.bootYX,typeBCa=FALSE)
plots.confints.bootpls(temp.ci)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
temp.ci <- confints.bootpls(Cornell.bootYX,typeBCa=FALSE)
plots.confints.bootpls(temp.ci)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
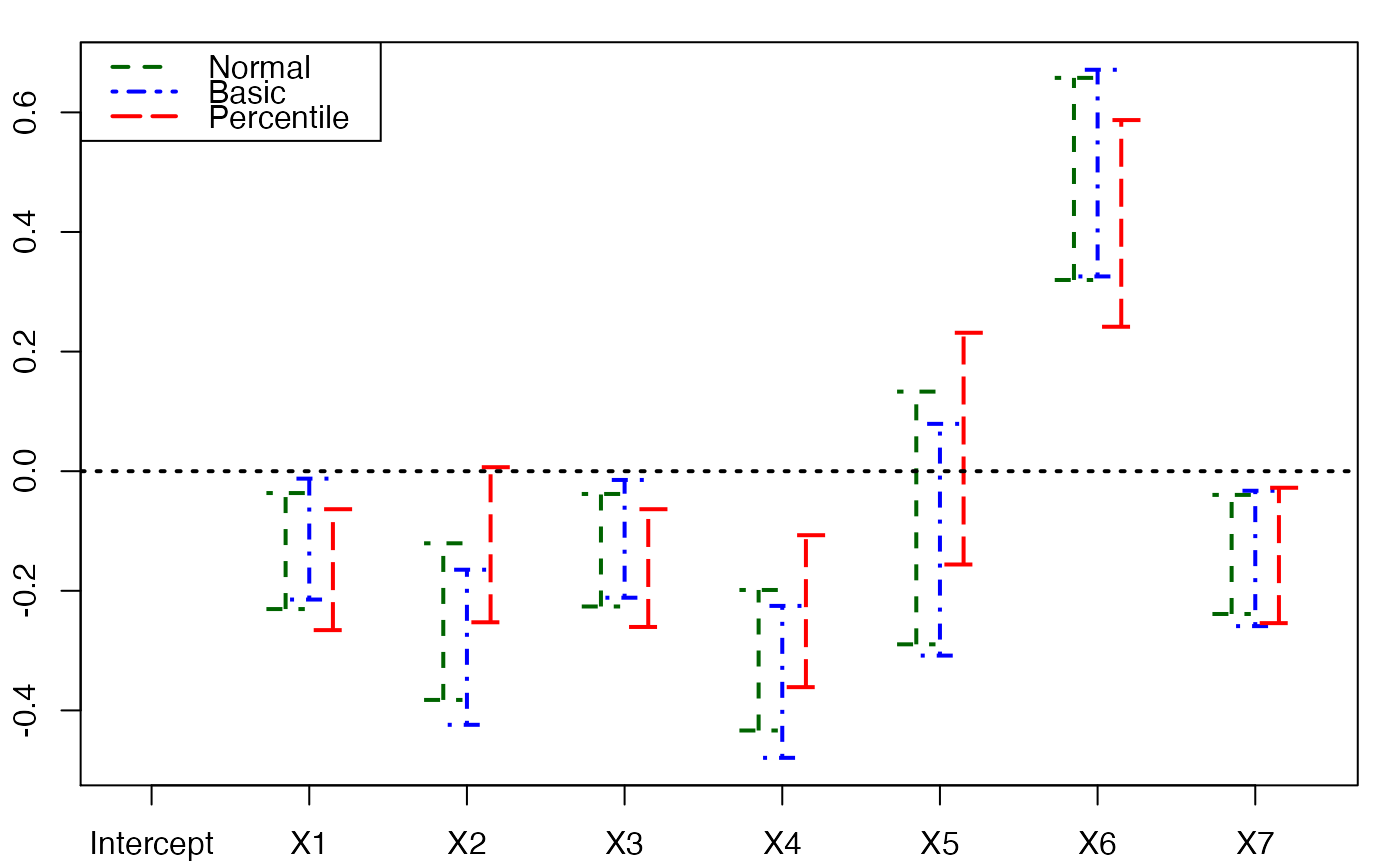 plots.confints.bootpls(temp.ci,2:8)
plots.confints.bootpls(temp.ci,2:8)
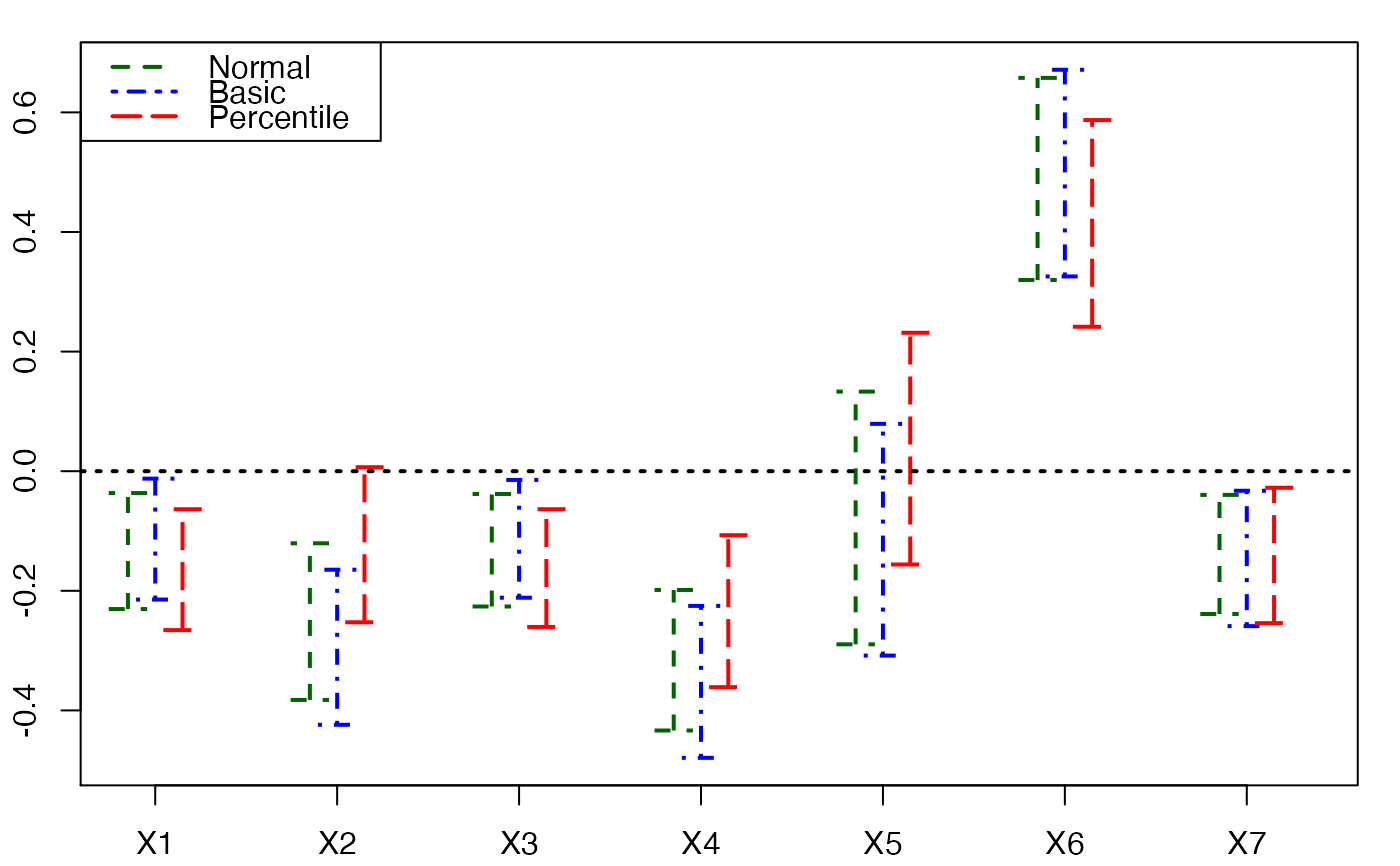 plots.confints.bootpls(temp.ci,prednames=FALSE)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
plots.confints.bootpls(temp.ci,prednames=FALSE)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
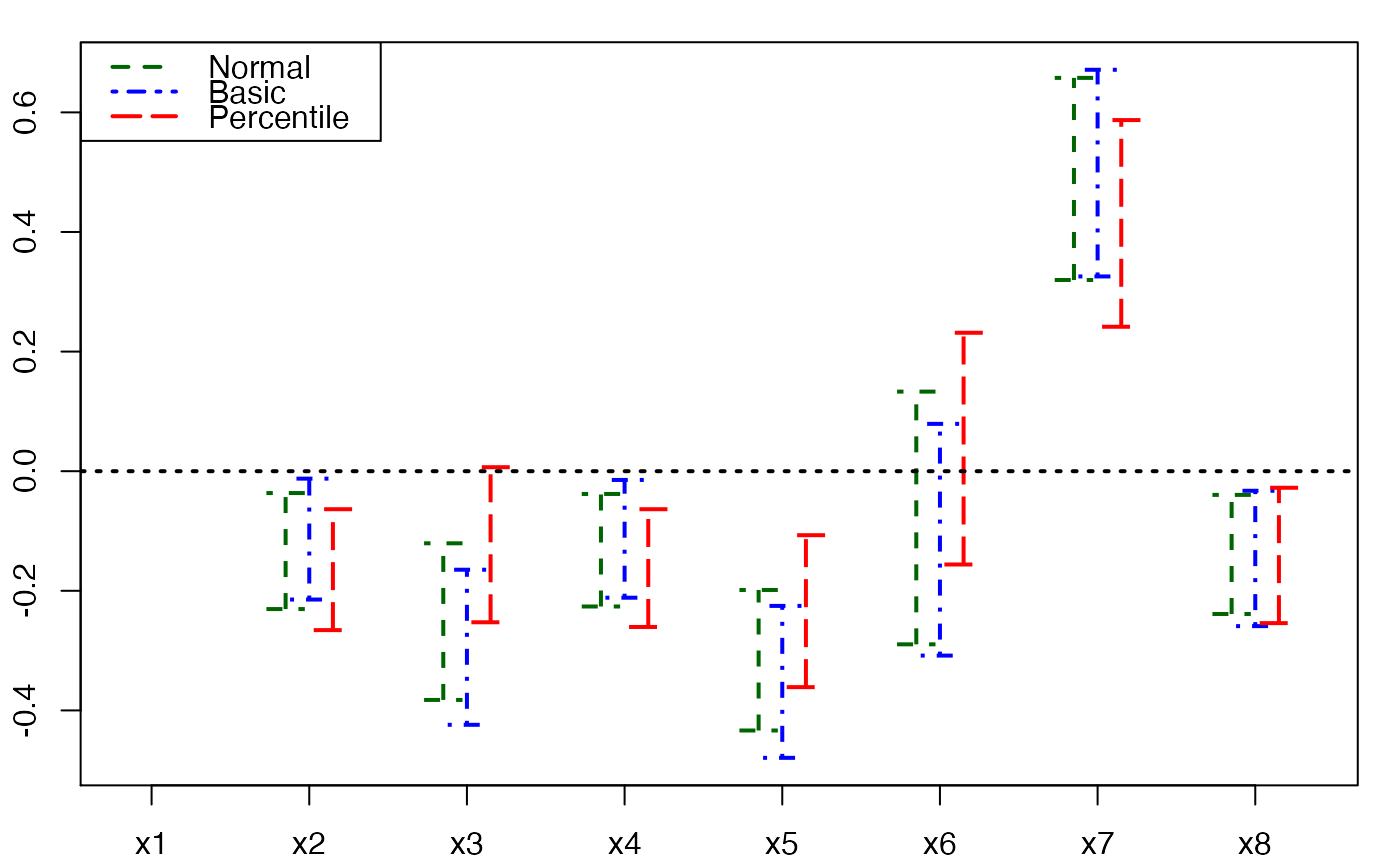 # Bastien CSDA 2005 (Y,T) bootstrap
Cornell.boot <- bootpls(modpls, typeboot="fmodel_np", R=250, verbose=FALSE)
temp.ci <- confints.bootpls(Cornell.boot,2:8)
plots.confints.bootpls(temp.ci)
# Bastien CSDA 2005 (Y,T) bootstrap
Cornell.boot <- bootpls(modpls, typeboot="fmodel_np", R=250, verbose=FALSE)
temp.ci <- confints.bootpls(Cornell.boot,2:8)
plots.confints.bootpls(temp.ci)
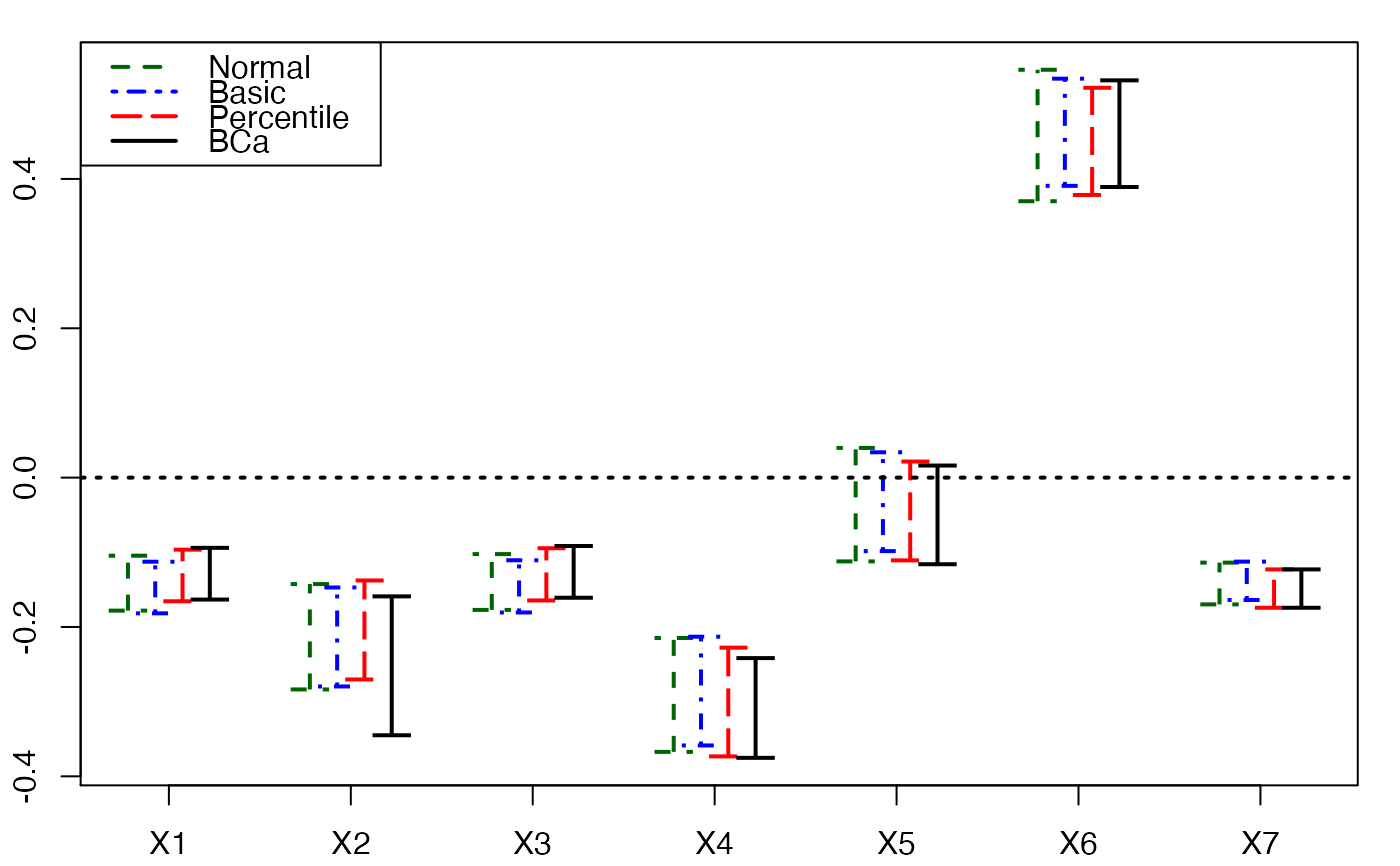 plots.confints.bootpls(temp.ci,prednames=FALSE)
plots.confints.bootpls(temp.ci,prednames=FALSE)
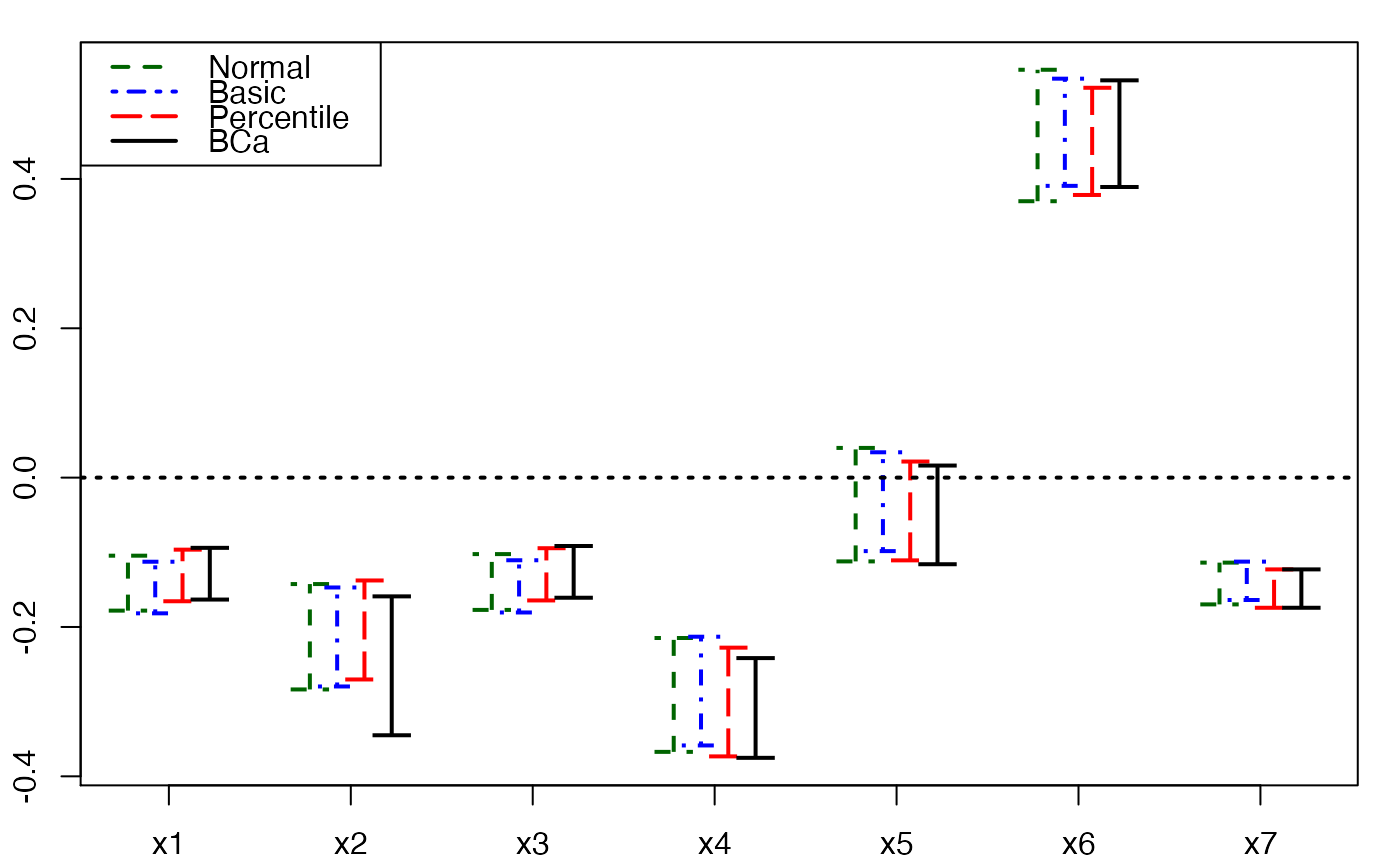 plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
 plots.confints.bootpls(temp.ci,indices=1:3,prednames=FALSE)
plots.confints.bootpls(temp.ci,indices=1:3,prednames=FALSE)
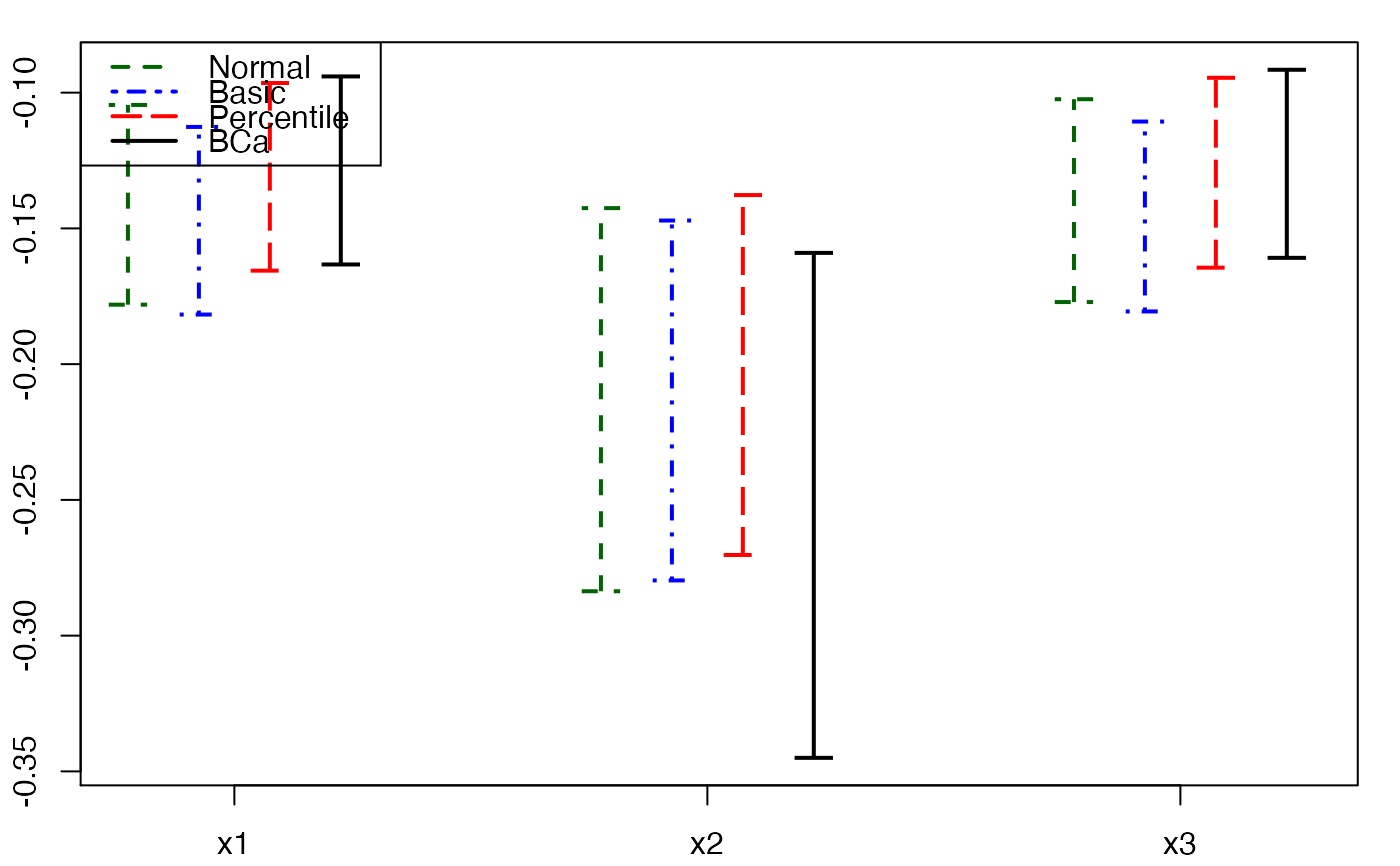 plots.confints.bootpls(temp.ci,c(2,4,6),"bottomright")
plots.confints.bootpls(temp.ci,c(2,4,6),"bottomright")
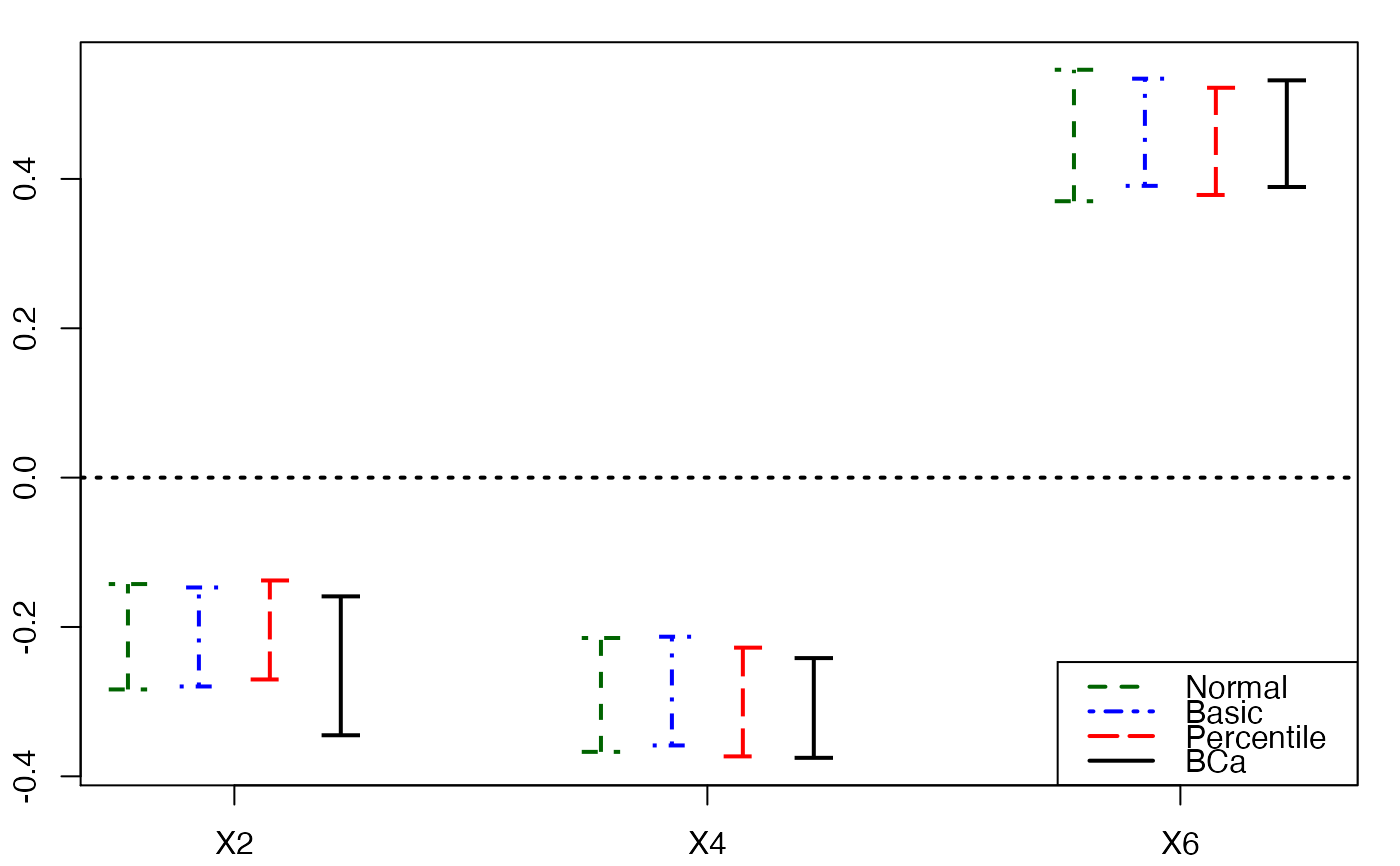 plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
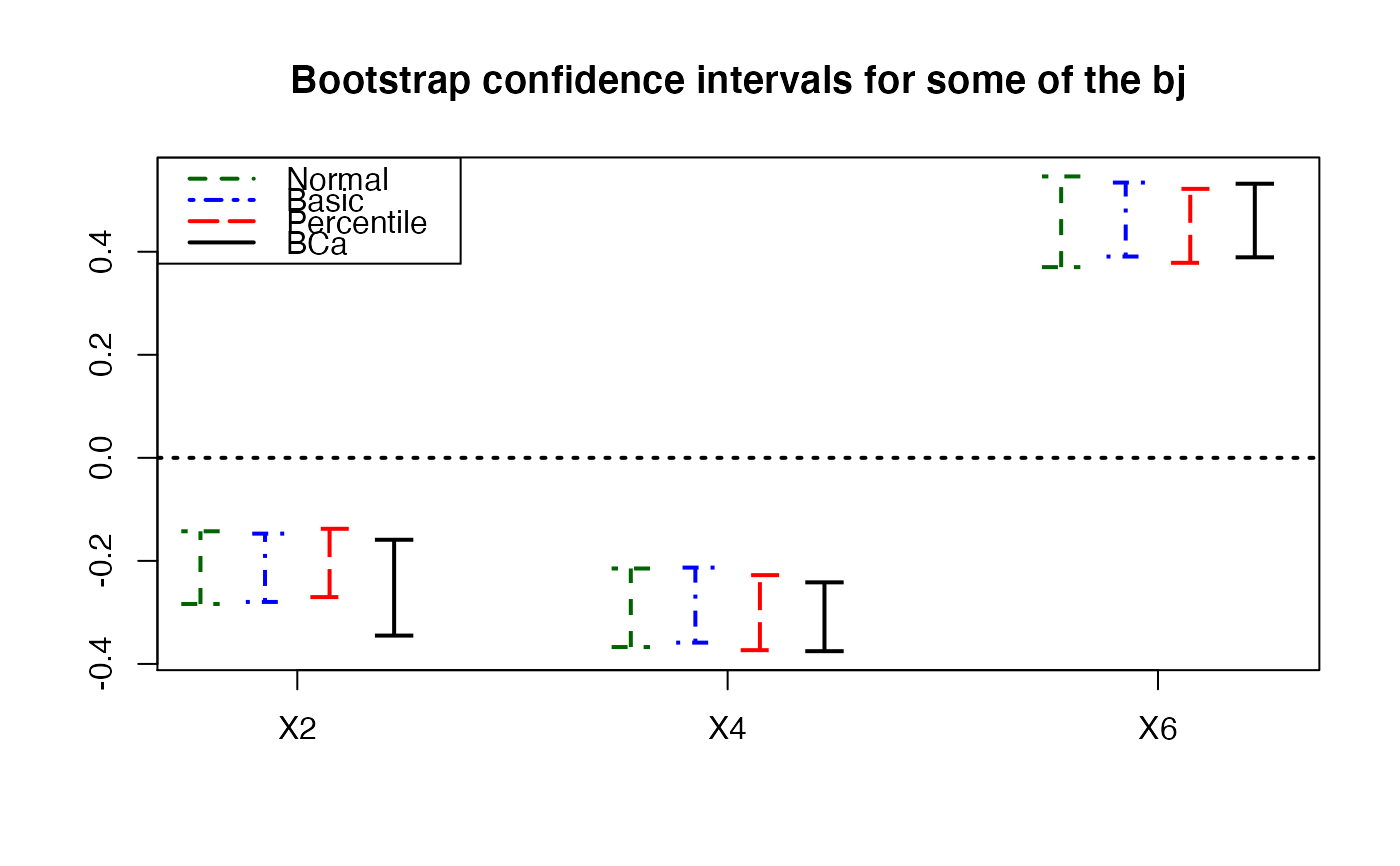
plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
 temp.ci <- confints.bootpls(Cornell.boot,typeBCa=FALSE)
plots.confints.bootpls(temp.ci)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
temp.ci <- confints.bootpls(Cornell.boot,typeBCa=FALSE)
plots.confints.bootpls(temp.ci)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
 plots.confints.bootpls(temp.ci,2:8)
plots.confints.bootpls(temp.ci,2:8)
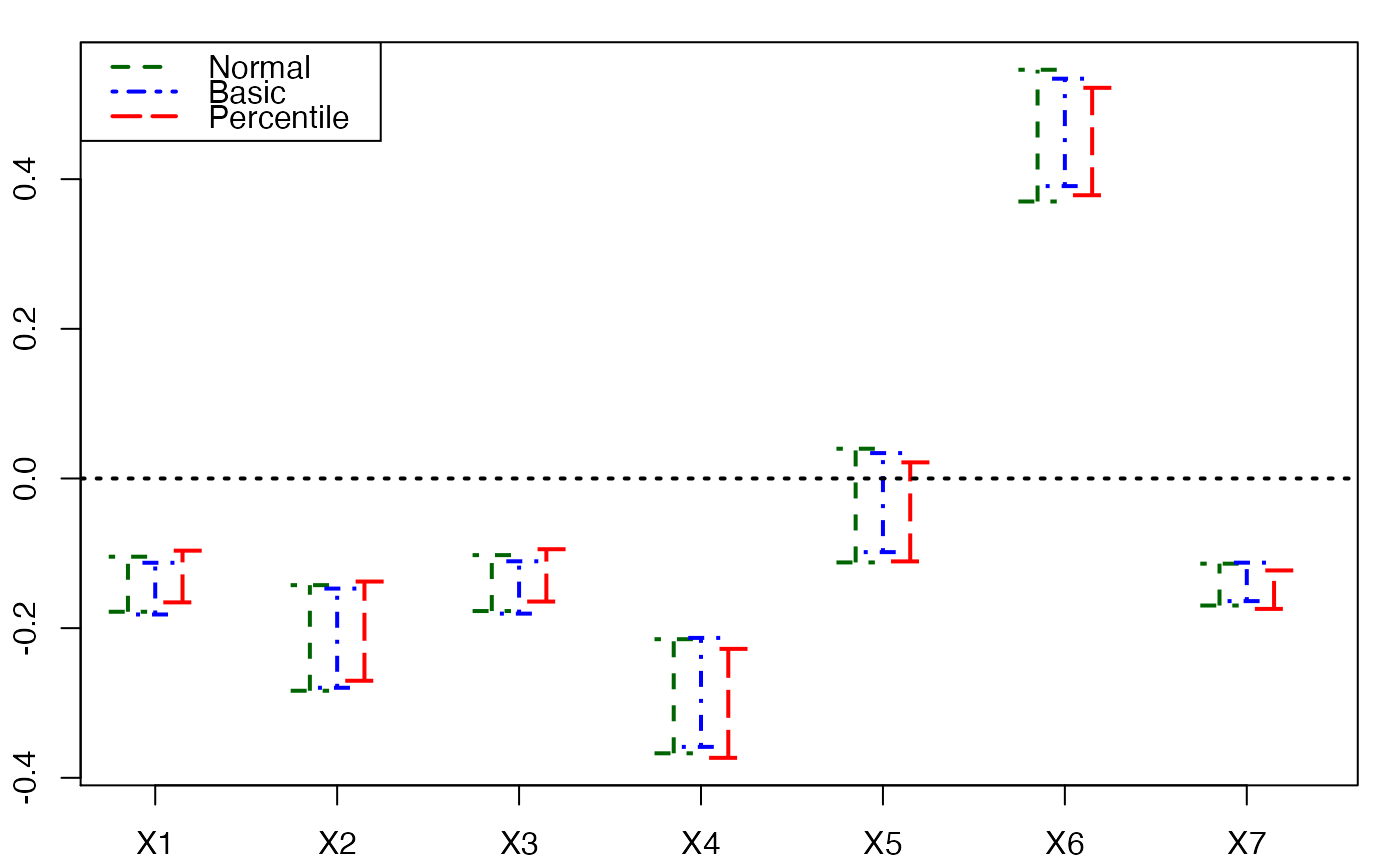 plots.confints.bootpls(temp.ci,prednames=FALSE)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
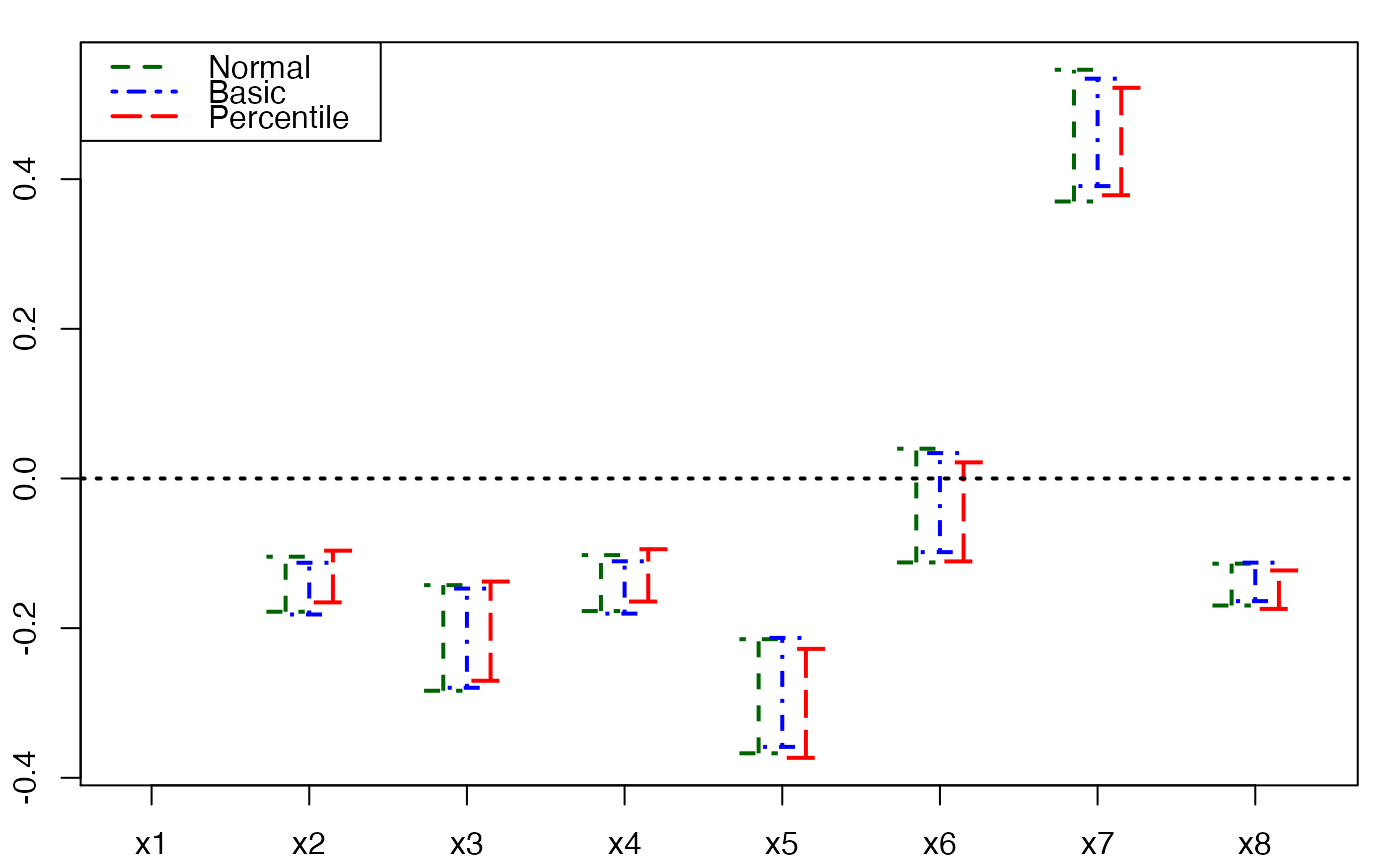
plots.confints.bootpls(temp.ci,prednames=FALSE)
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
#> Warning: zero-length arrow is of indeterminate angle and so skipped
 # \donttest{
data(aze_compl)
modplsglm <- plsRglm(y~.,data=aze_compl,3,modele="pls-glm-logistic")
#> ____************************************************____
#>
#> Family: binomial
#> Link function: logit
#>
#> ____Component____ 1 ____
#> ____Component____ 2 ____
#> ____Component____ 3 ____
#> ____Predicting X without NA neither in X or Y____
#> ****________________________________________________****
#>
# Lazraq-Cleroux PLS (Y,X) bootstrap
# should be run with R=1000 but takes much longer time
aze_compl.bootYX3 <- bootplsglm(modplsglm, typeboot="plsmodel", R=250, verbose=FALSE)
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
temp.ci <- confints.bootpls(aze_compl.bootYX3)
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
plots.confints.bootpls(temp.ci)
# \donttest{
data(aze_compl)
modplsglm <- plsRglm(y~.,data=aze_compl,3,modele="pls-glm-logistic")
#> ____************************************************____
#>
#> Family: binomial
#> Link function: logit
#>
#> ____Component____ 1 ____
#> ____Component____ 2 ____
#> ____Component____ 3 ____
#> ____Predicting X without NA neither in X or Y____
#> ****________________________________________________****
#>
# Lazraq-Cleroux PLS (Y,X) bootstrap
# should be run with R=1000 but takes much longer time
aze_compl.bootYX3 <- bootplsglm(modplsglm, typeboot="plsmodel", R=250, verbose=FALSE)
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
#> Warning: glm.fit: fitted probabilities numerically 0 or 1 occurred
temp.ci <- confints.bootpls(aze_compl.bootYX3)
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
#> Warning: extreme order statistics used as endpoints
plots.confints.bootpls(temp.ci)
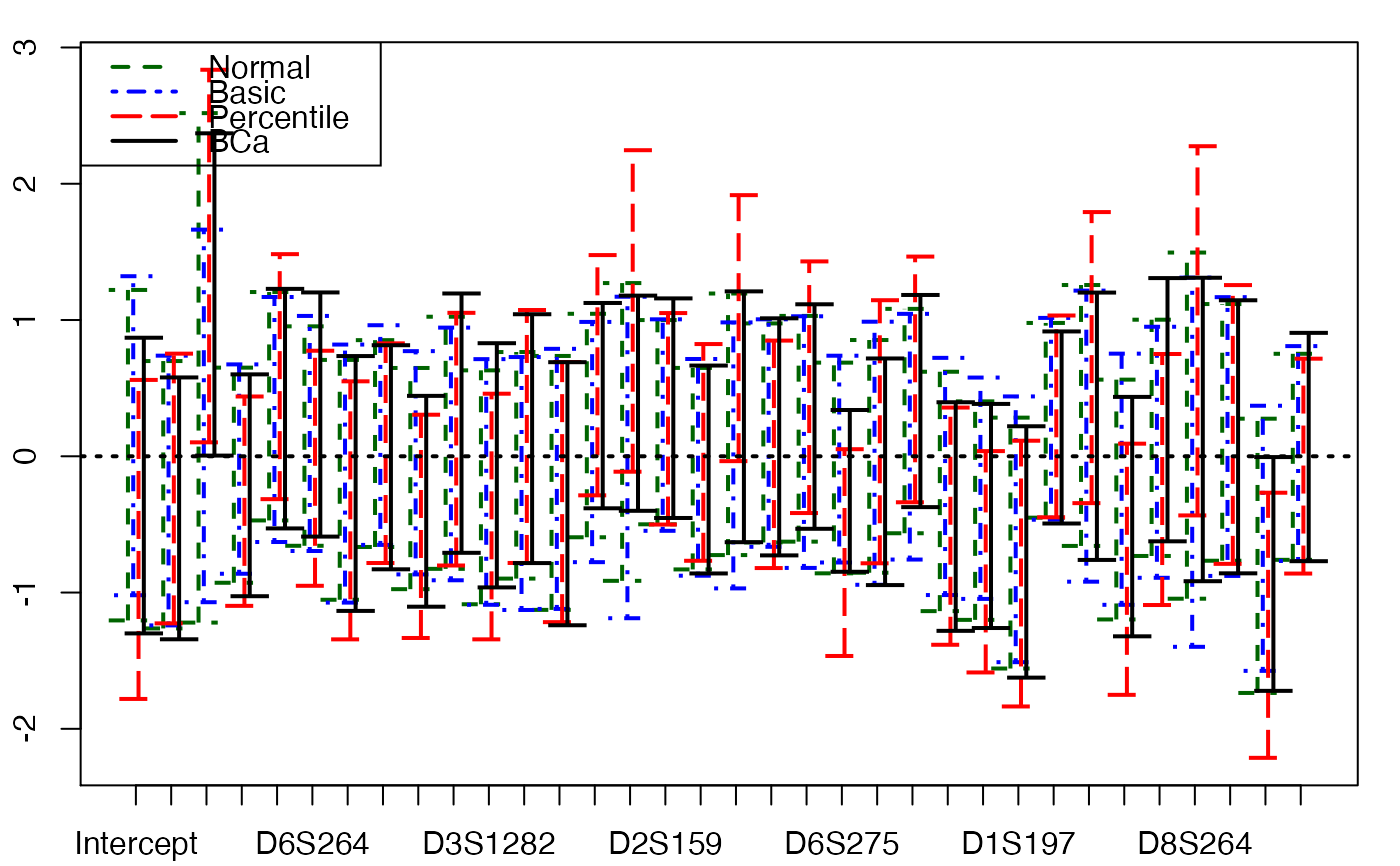 plots.confints.bootpls(temp.ci,prednames=FALSE)
plots.confints.bootpls(temp.ci,prednames=FALSE)
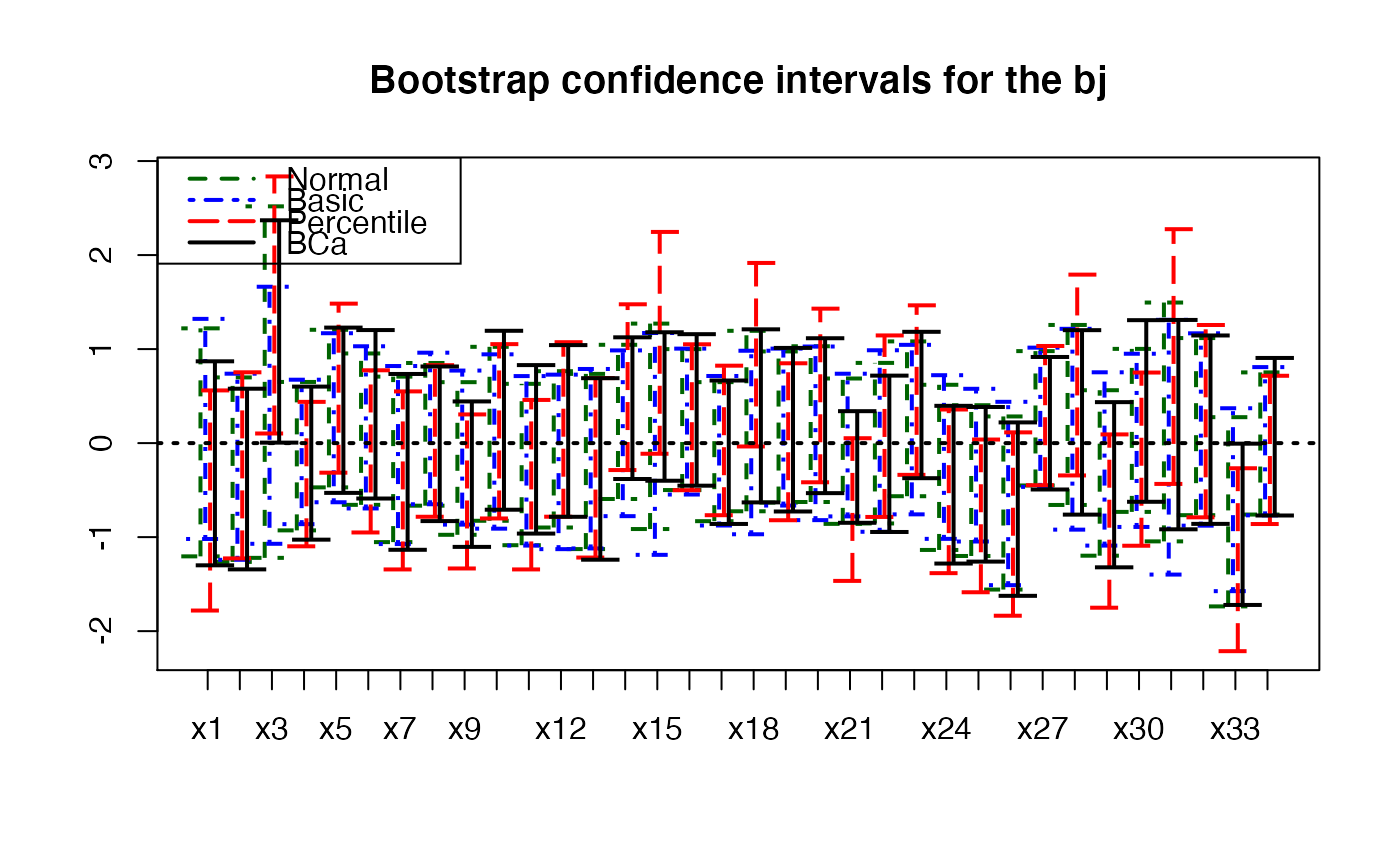
 plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
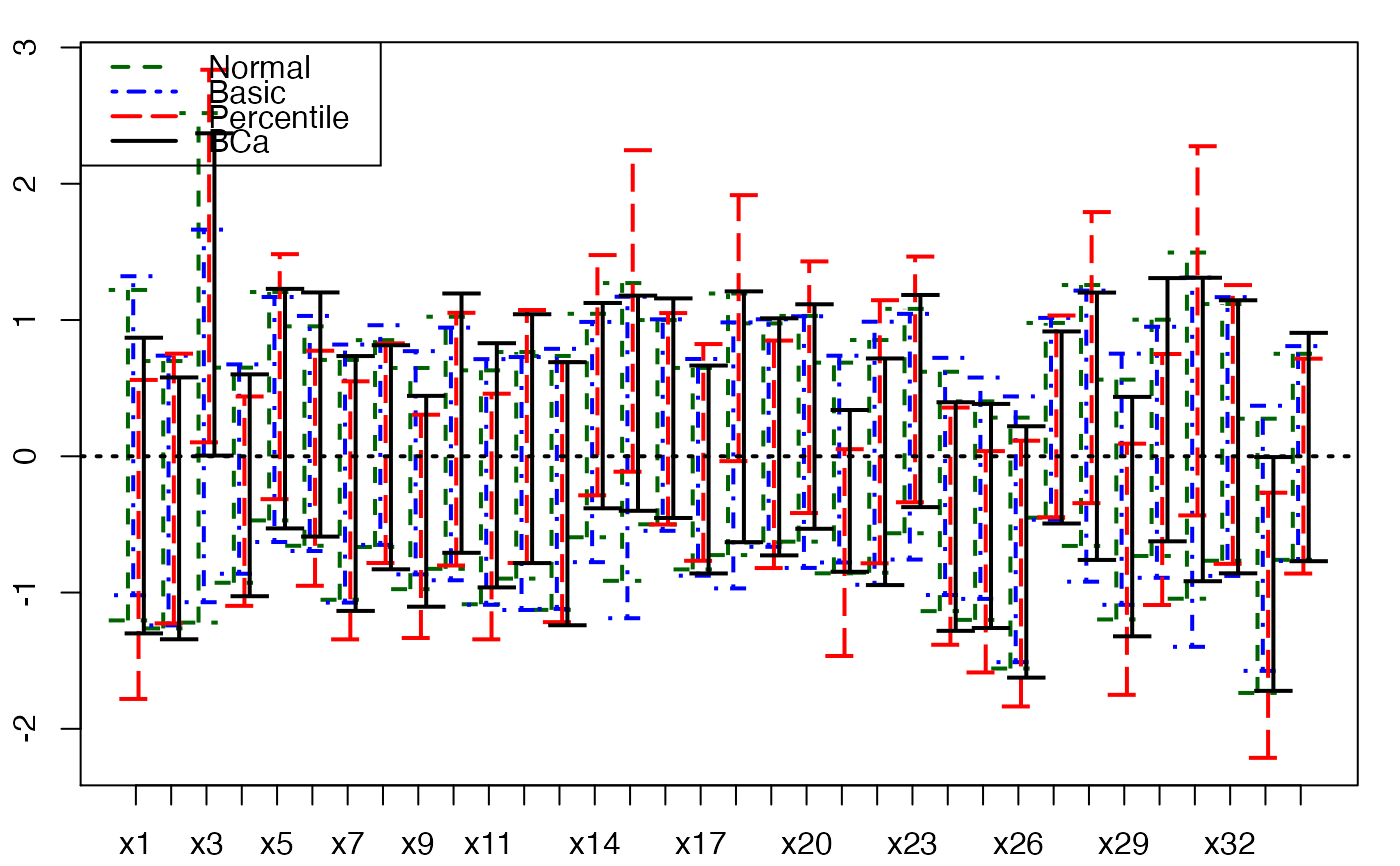
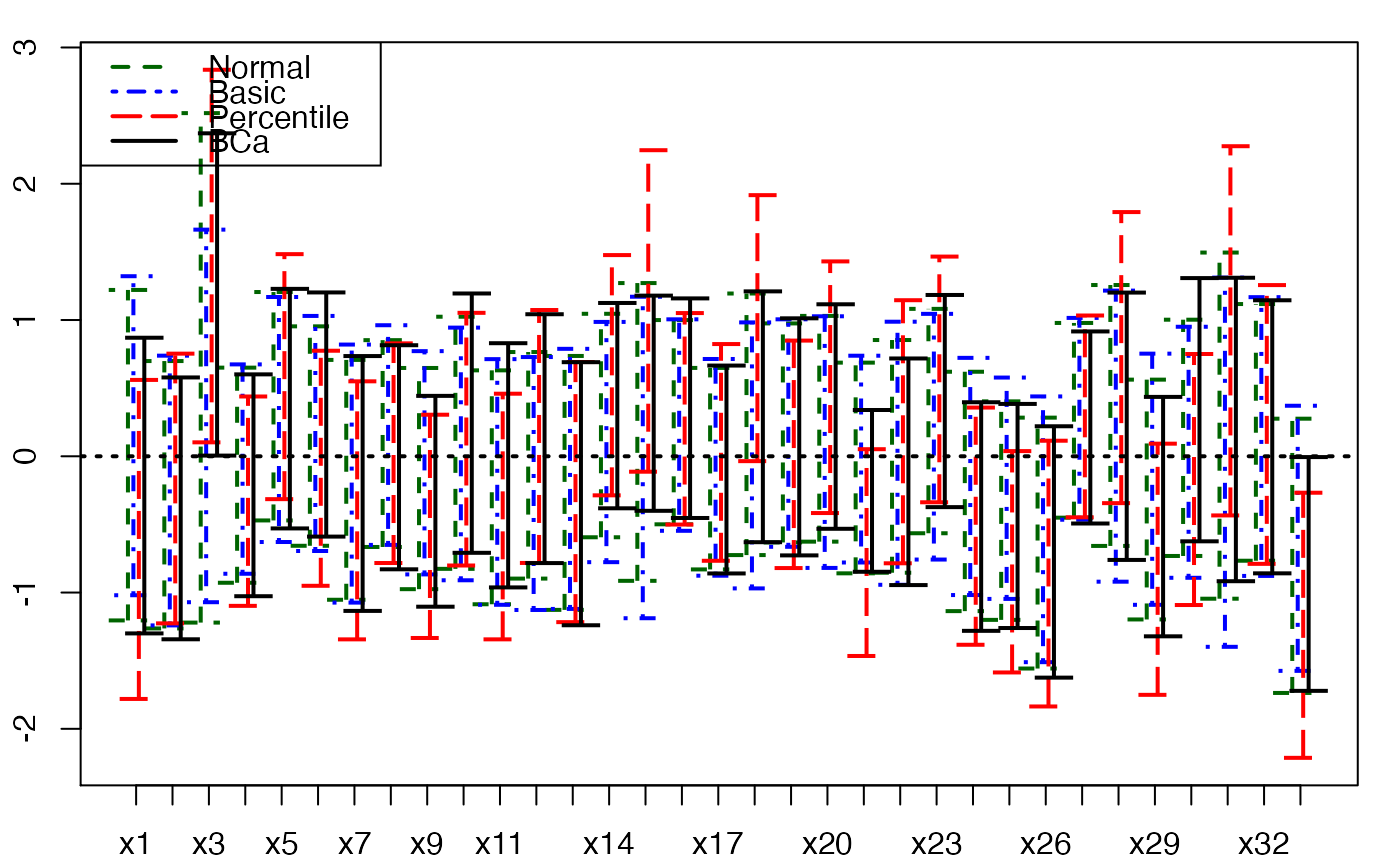
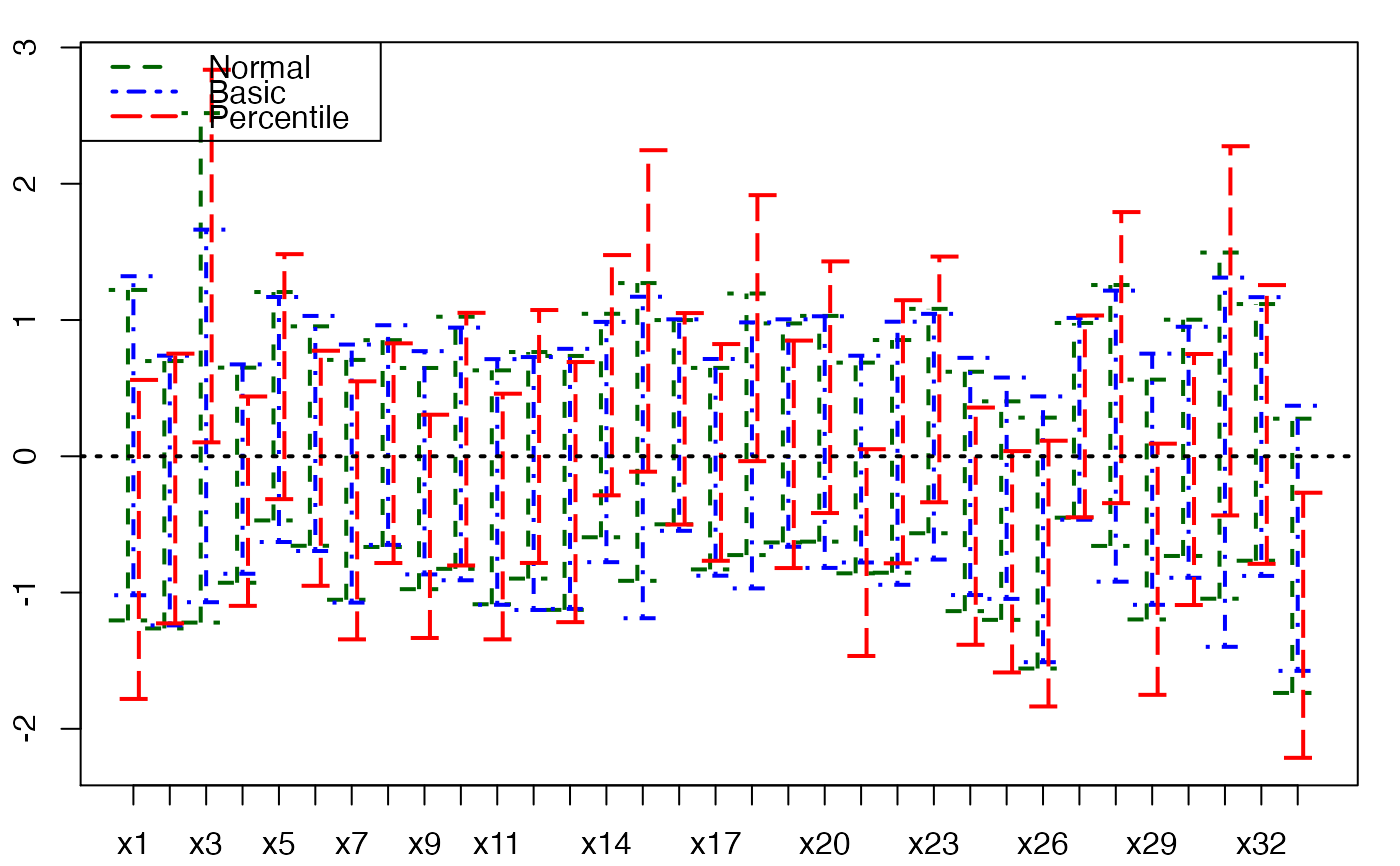
 plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE)
plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE)
 plots.confints.bootpls(temp.ci,c(2,4,6),"bottomleft")
plots.confints.bootpls(temp.ci,c(2,4,6),"bottomleft")
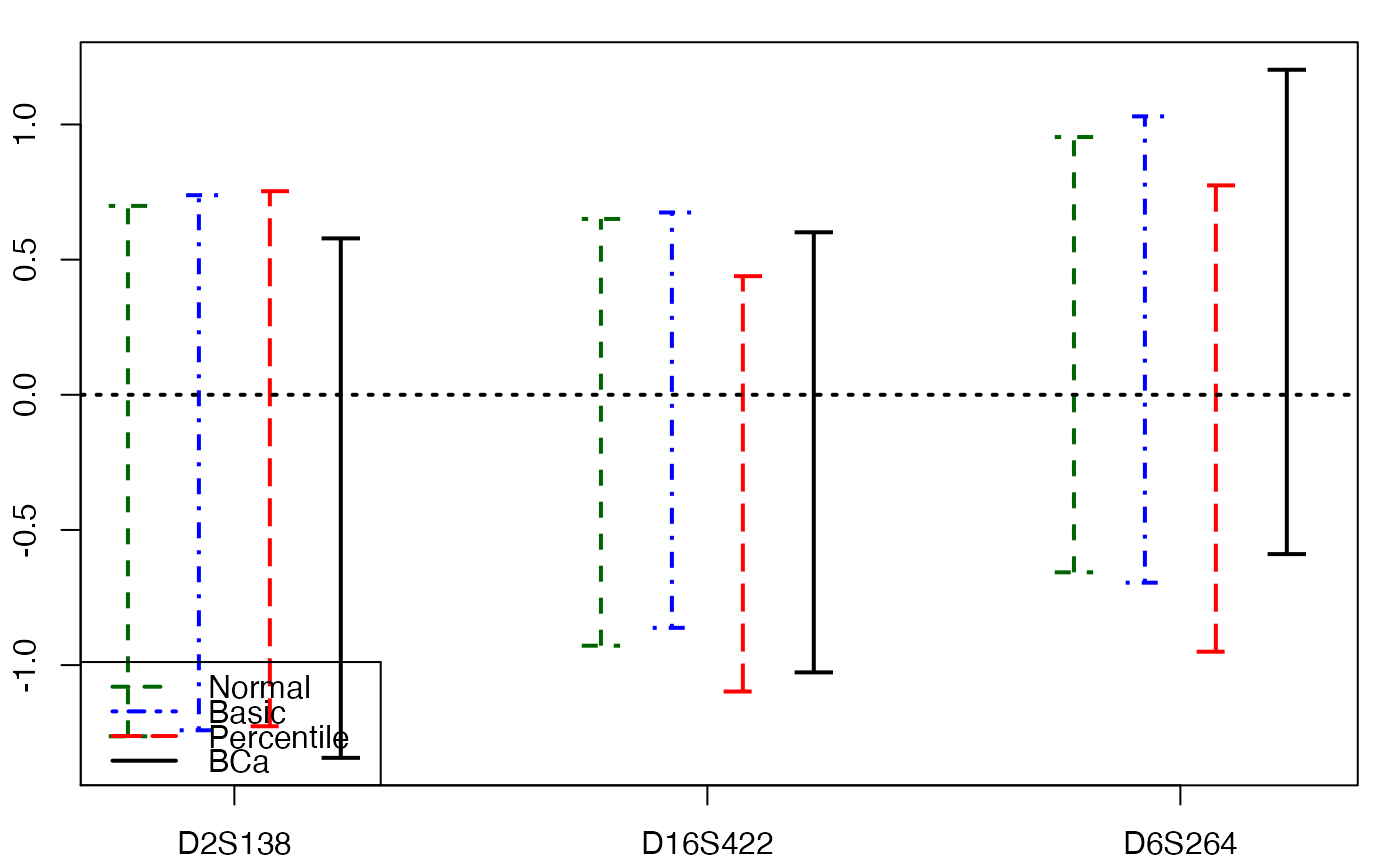 plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
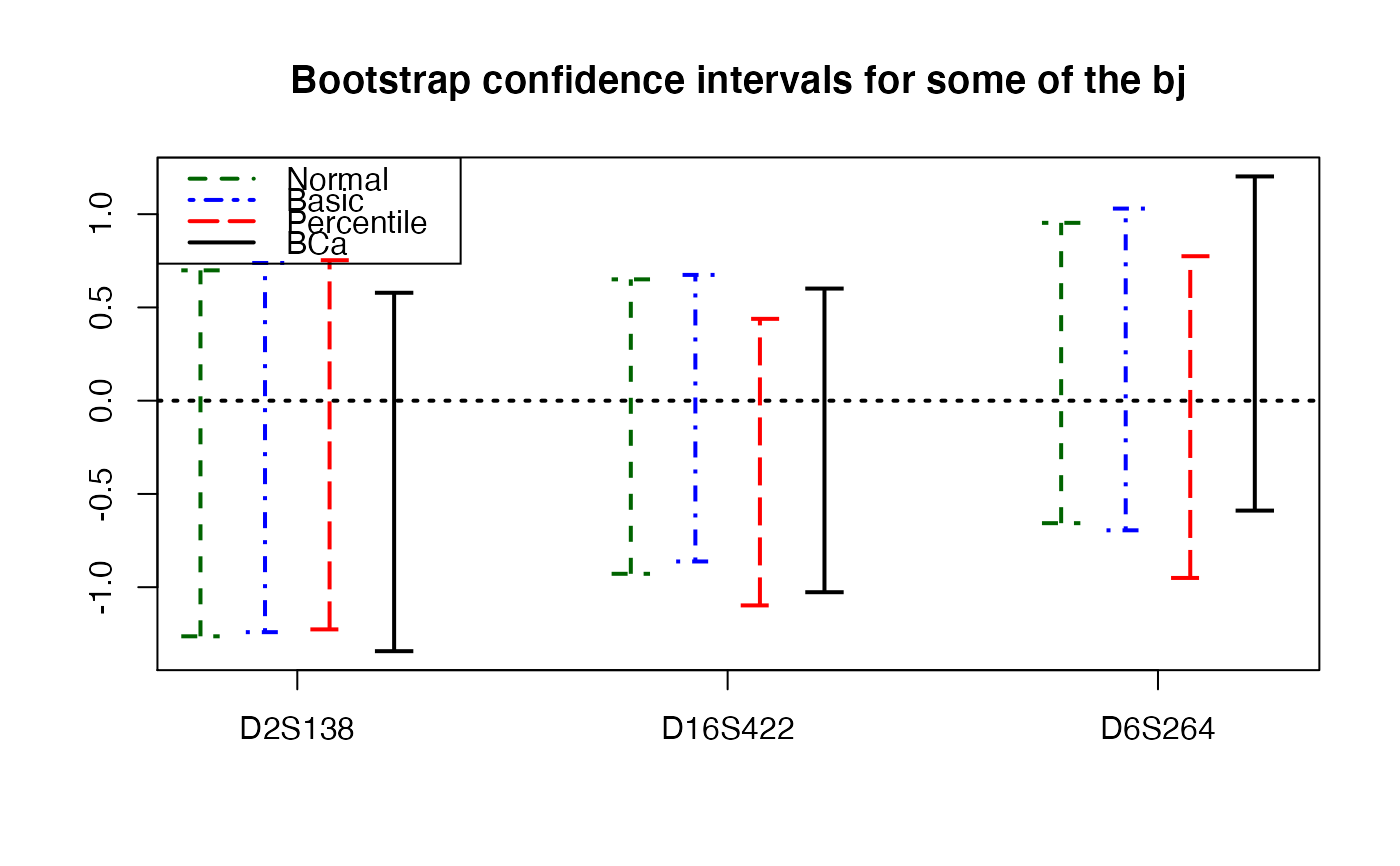 plots.confints.bootpls(temp.ci,indices=1:34,prednames=FALSE)
plots.confints.bootpls(temp.ci,indices=1:34,prednames=FALSE)
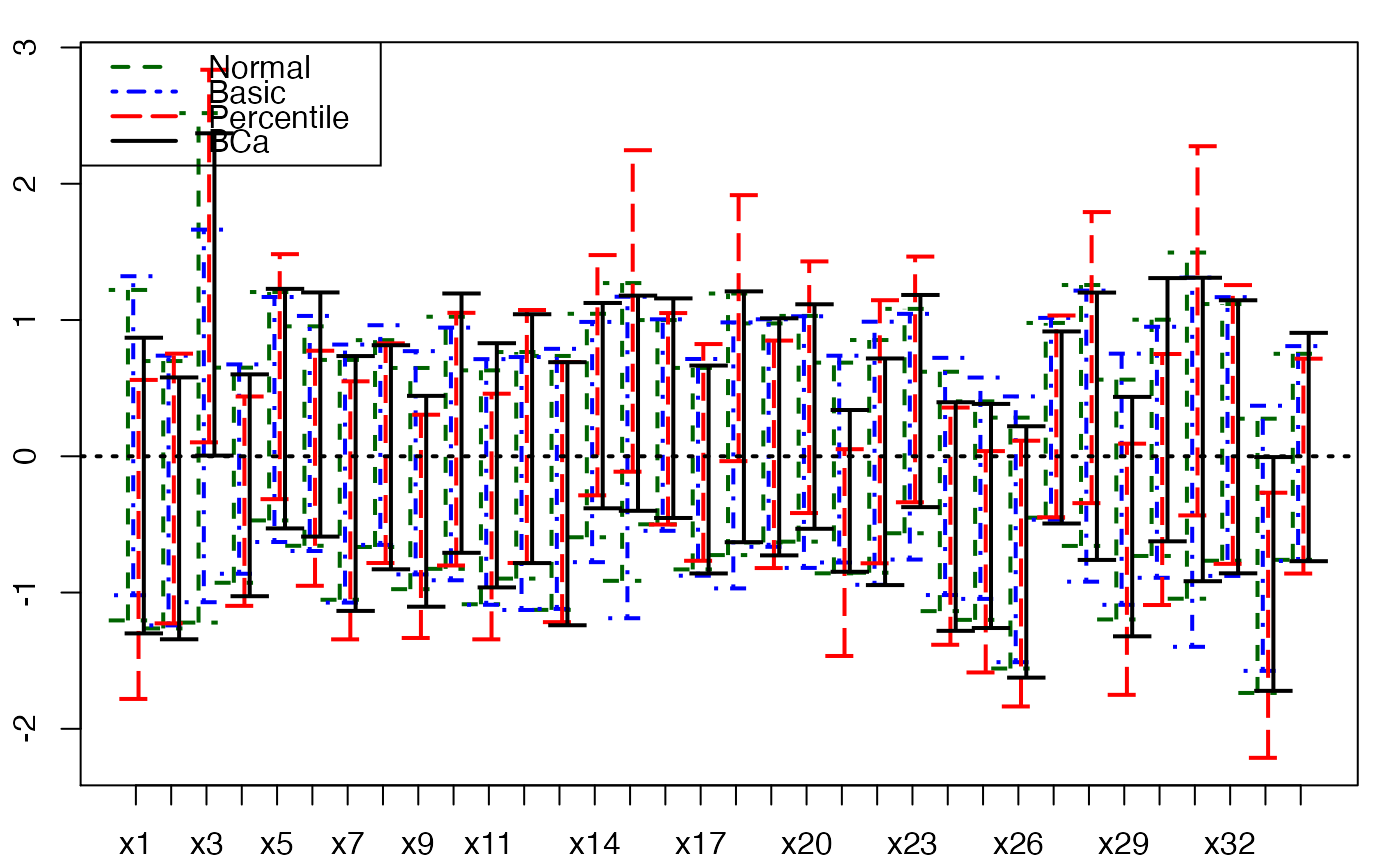 plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE,ltyIC=1,colIC=c(1,2))
plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE,ltyIC=1,colIC=c(1,2))
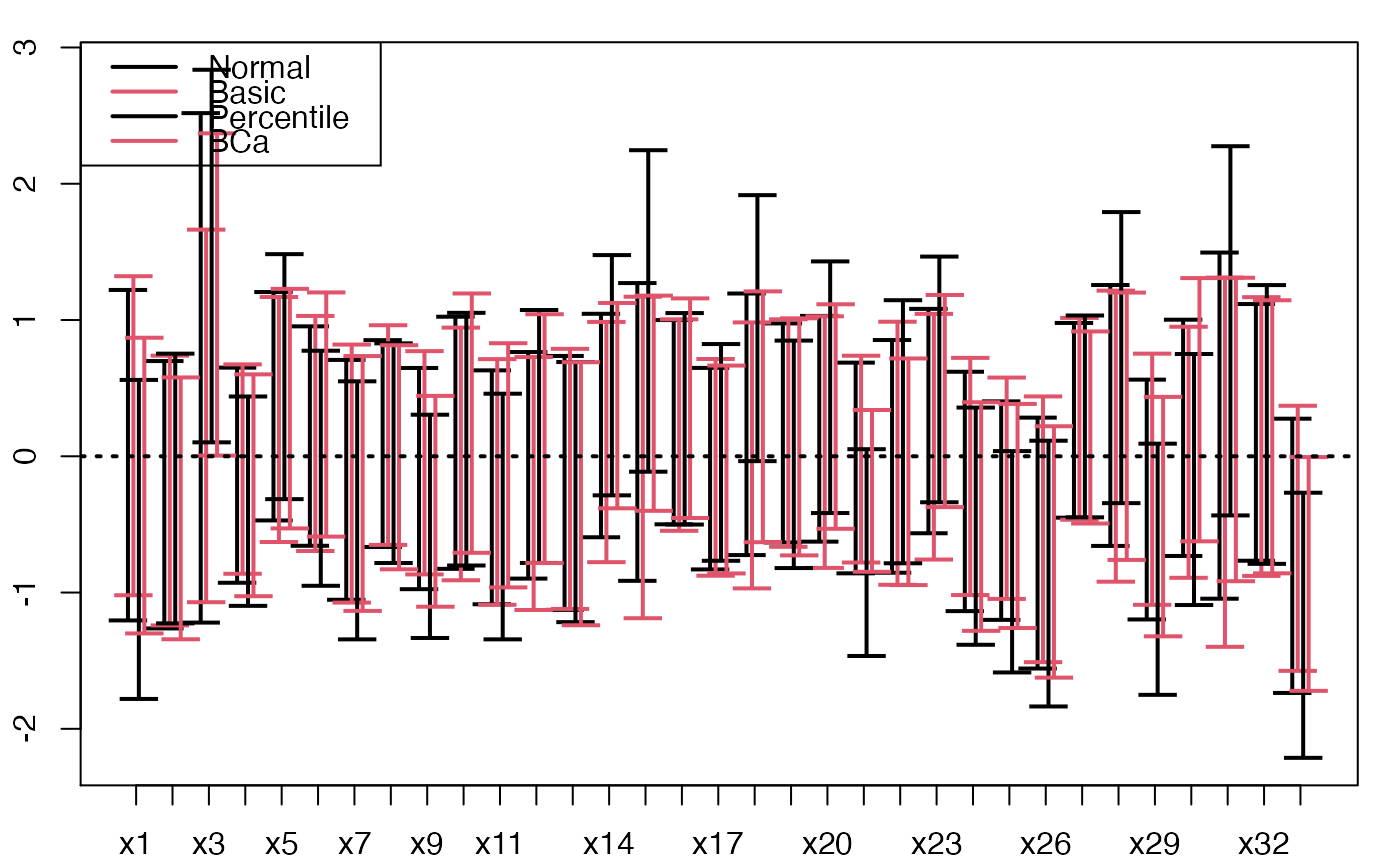 temp.ci <- confints.bootpls(aze_compl.bootYX3,1:34,typeBCa=FALSE)
plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE)
temp.ci <- confints.bootpls(aze_compl.bootYX3,1:34,typeBCa=FALSE)
plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE)
 # Bastien CSDA 2005 (Y,T) Bootstrap
# much faster
aze_compl.bootYT3 <- bootplsglm(modplsglm, R=1000, verbose=FALSE)
temp.ci <- confints.bootpls(aze_compl.bootYT3)
plots.confints.bootpls(temp.ci)
# Bastien CSDA 2005 (Y,T) Bootstrap
# much faster
aze_compl.bootYT3 <- bootplsglm(modplsglm, R=1000, verbose=FALSE)
temp.ci <- confints.bootpls(aze_compl.bootYT3)
plots.confints.bootpls(temp.ci)
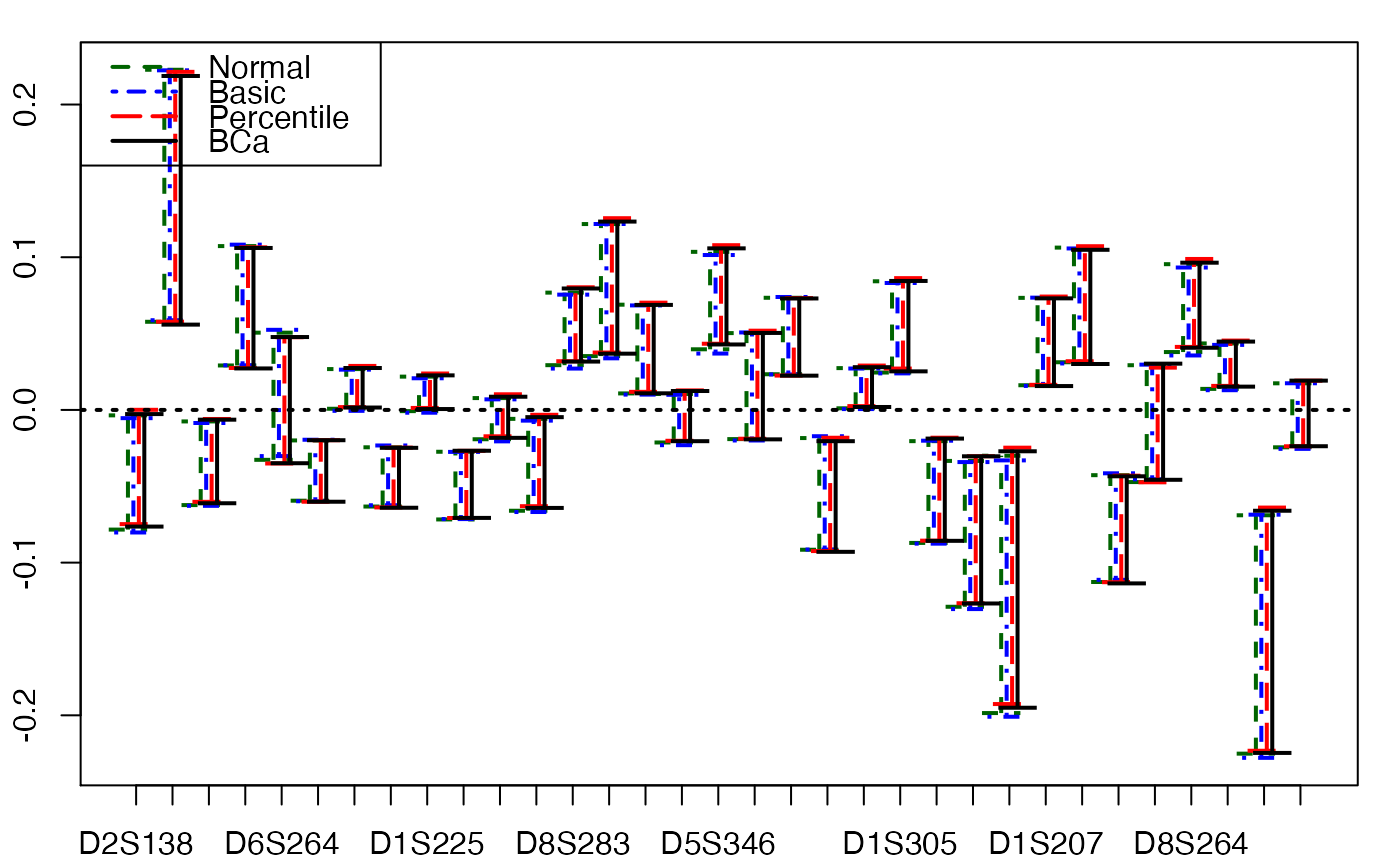 plots.confints.bootpls(temp.ci,typeIC="Normal")
plots.confints.bootpls(temp.ci,typeIC="Normal")
 plots.confints.bootpls(temp.ci,typeIC=c("Normal","Basic"))
plots.confints.bootpls(temp.ci,typeIC=c("Normal","Basic"))
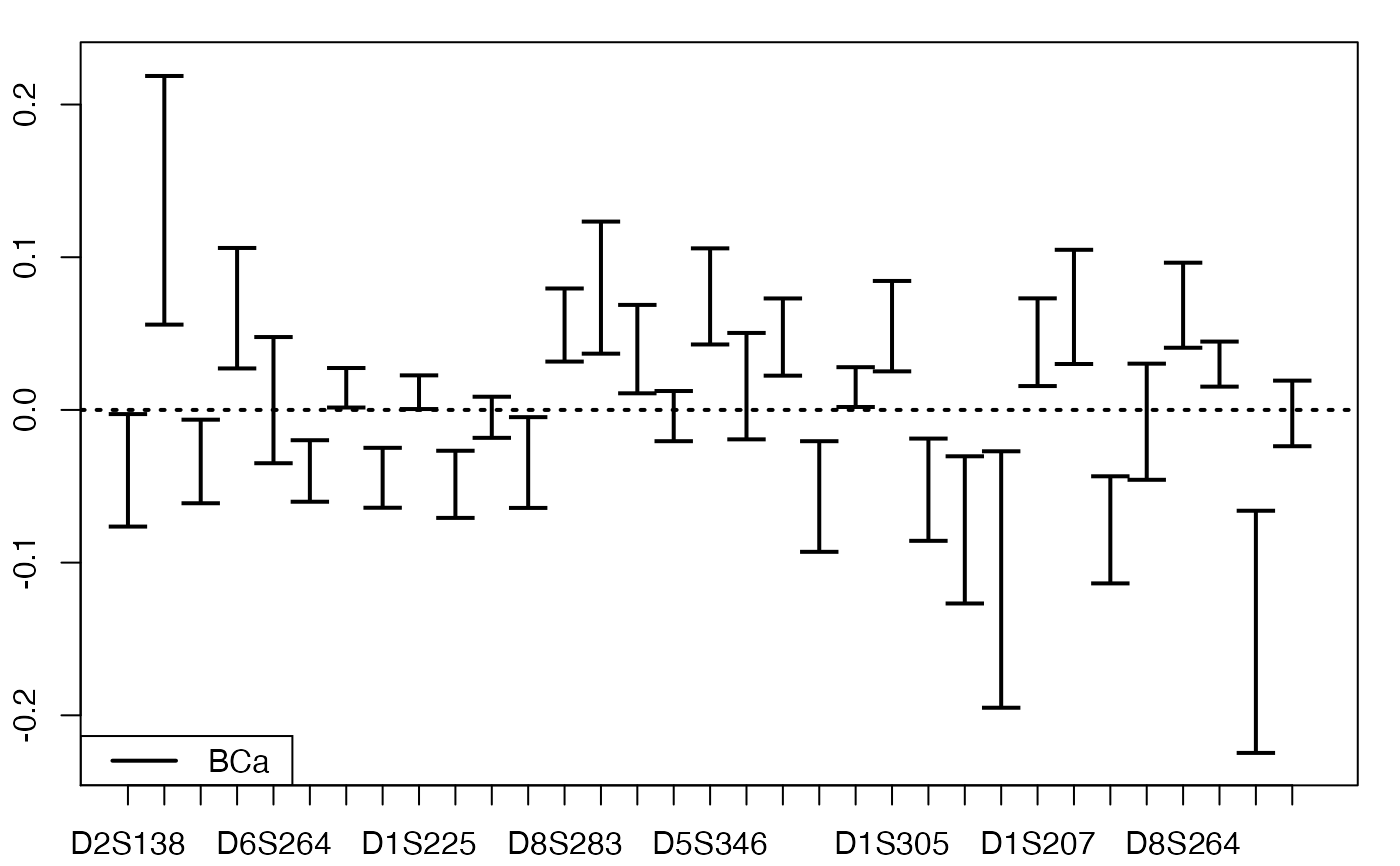
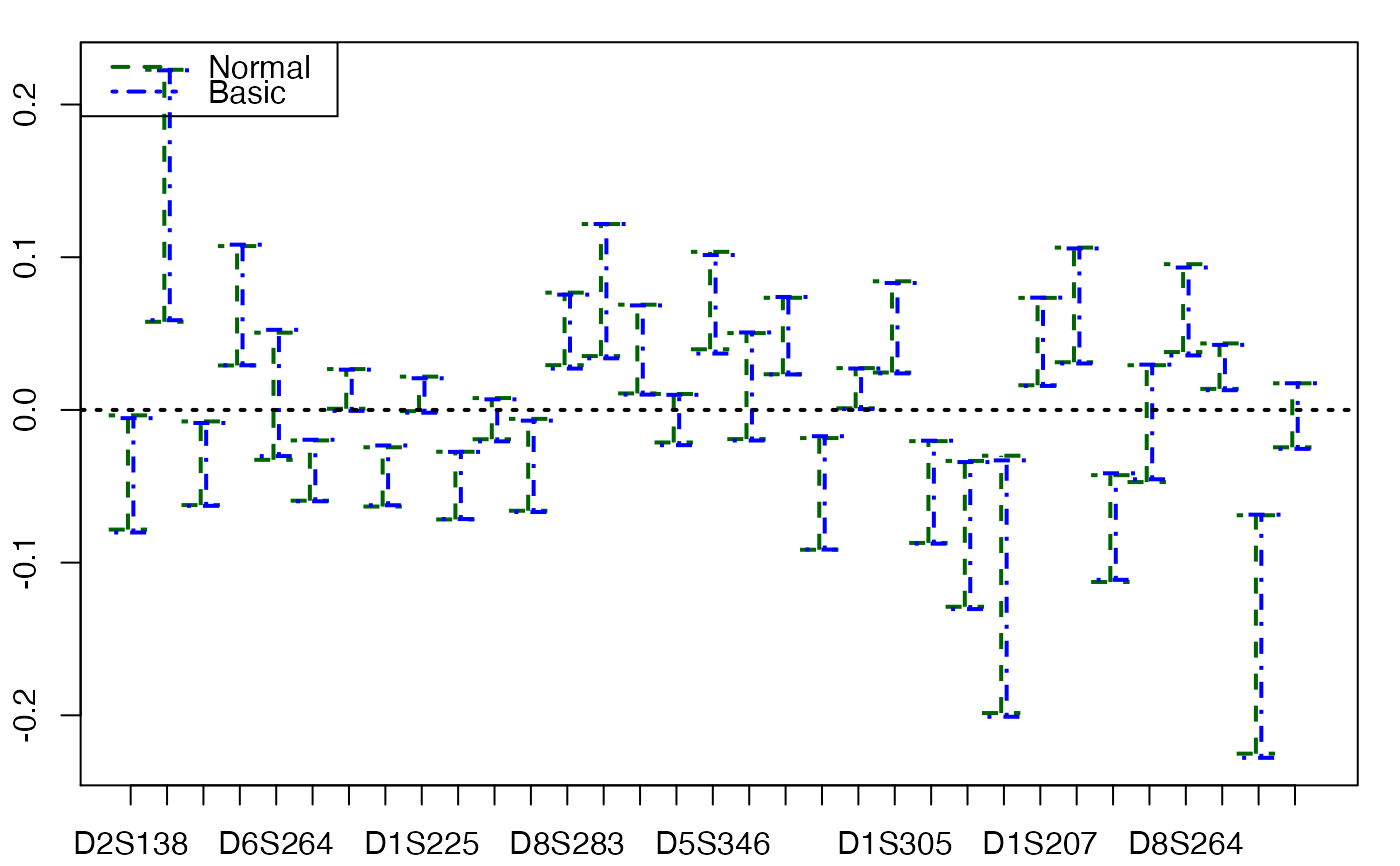 plots.confints.bootpls(temp.ci,typeIC="BCa",legendpos="bottomleft")
plots.confints.bootpls(temp.ci,typeIC="BCa",legendpos="bottomleft")
 plots.confints.bootpls(temp.ci,prednames=FALSE)
plots.confints.bootpls(temp.ci,prednames=FALSE)
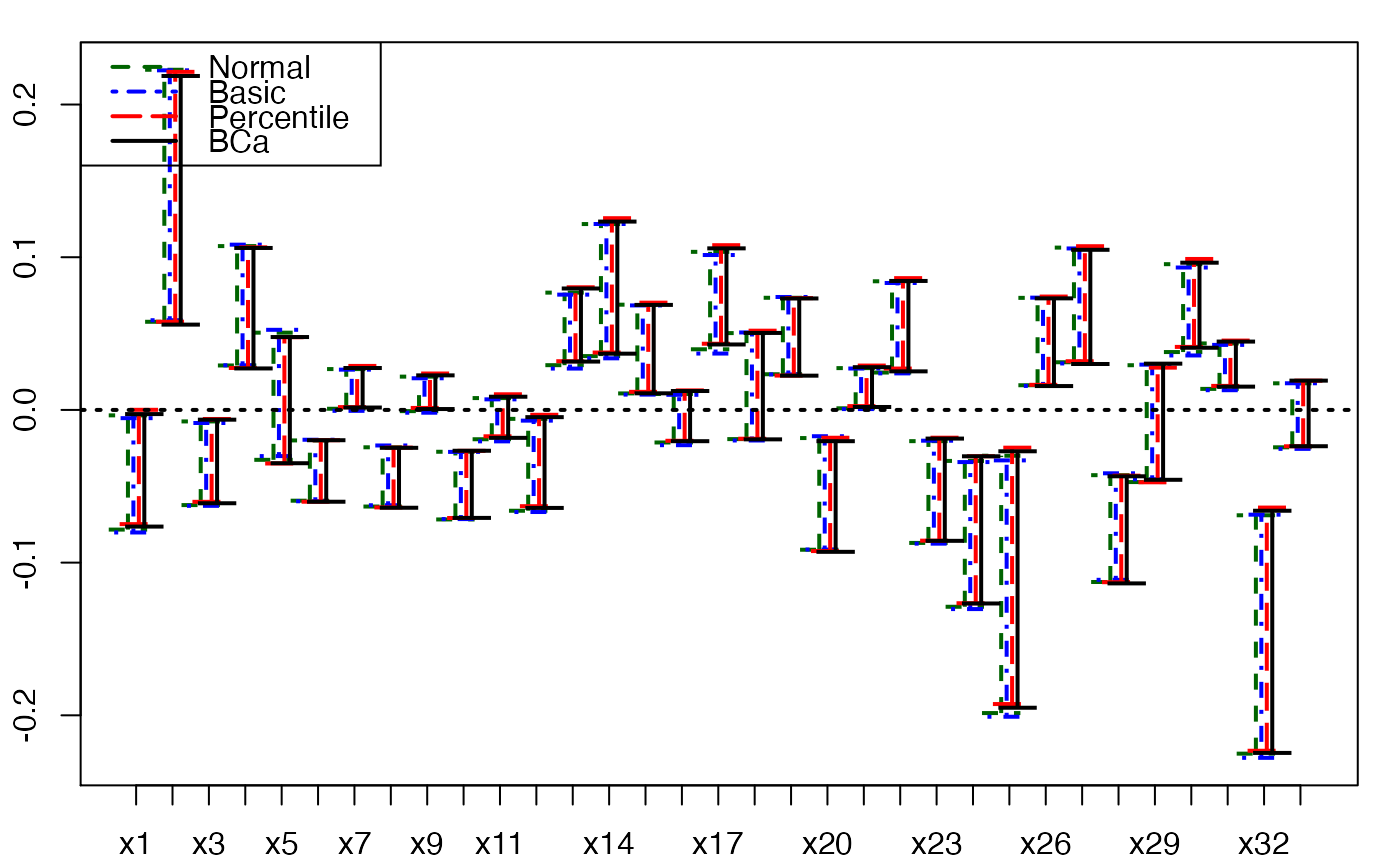 plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
plots.confints.bootpls(temp.ci,prednames=FALSE,articlestyle=FALSE,
main="Bootstrap confidence intervals for the bj")
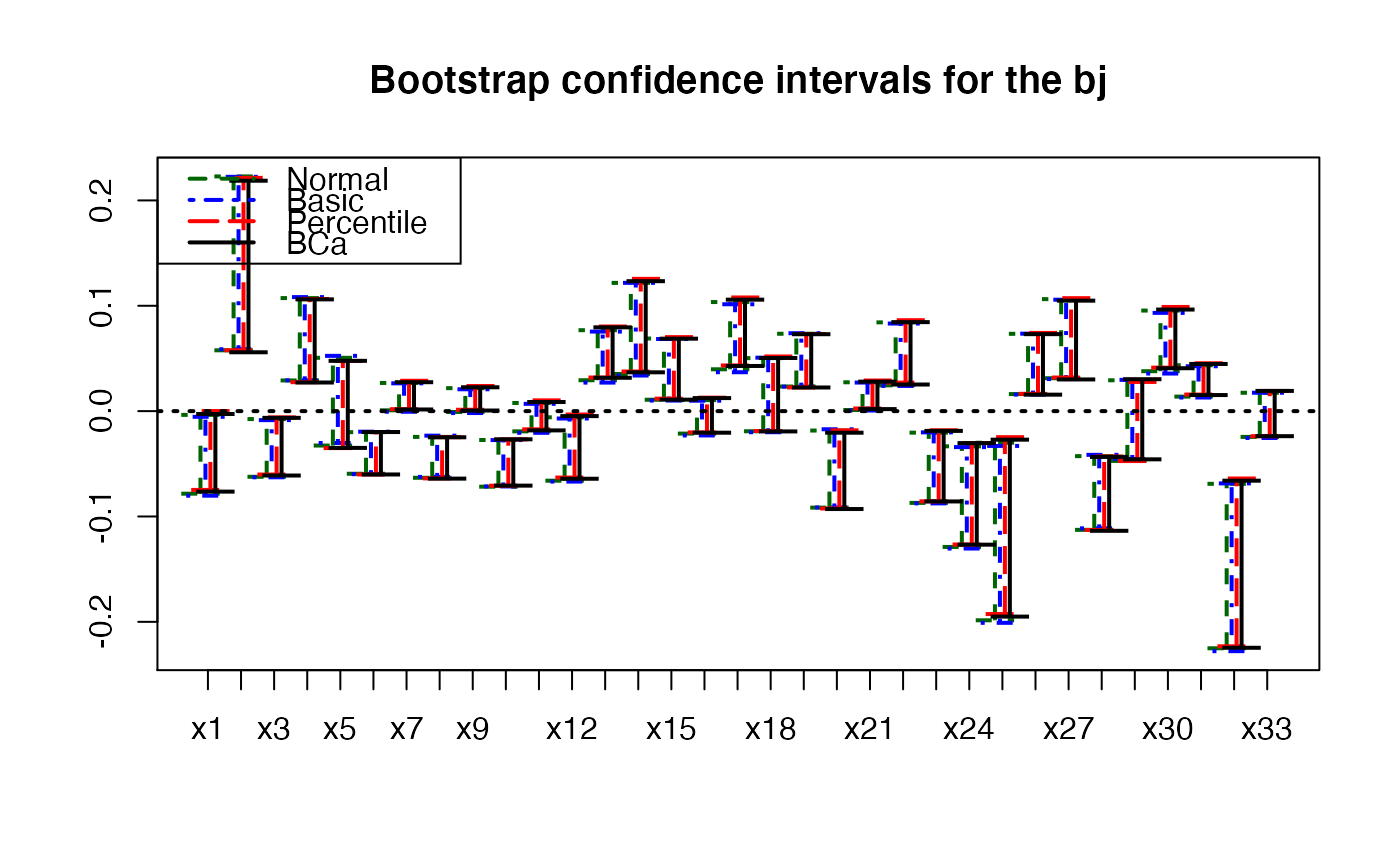 plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE)
plots.confints.bootpls(temp.ci,indices=1:33,prednames=FALSE)
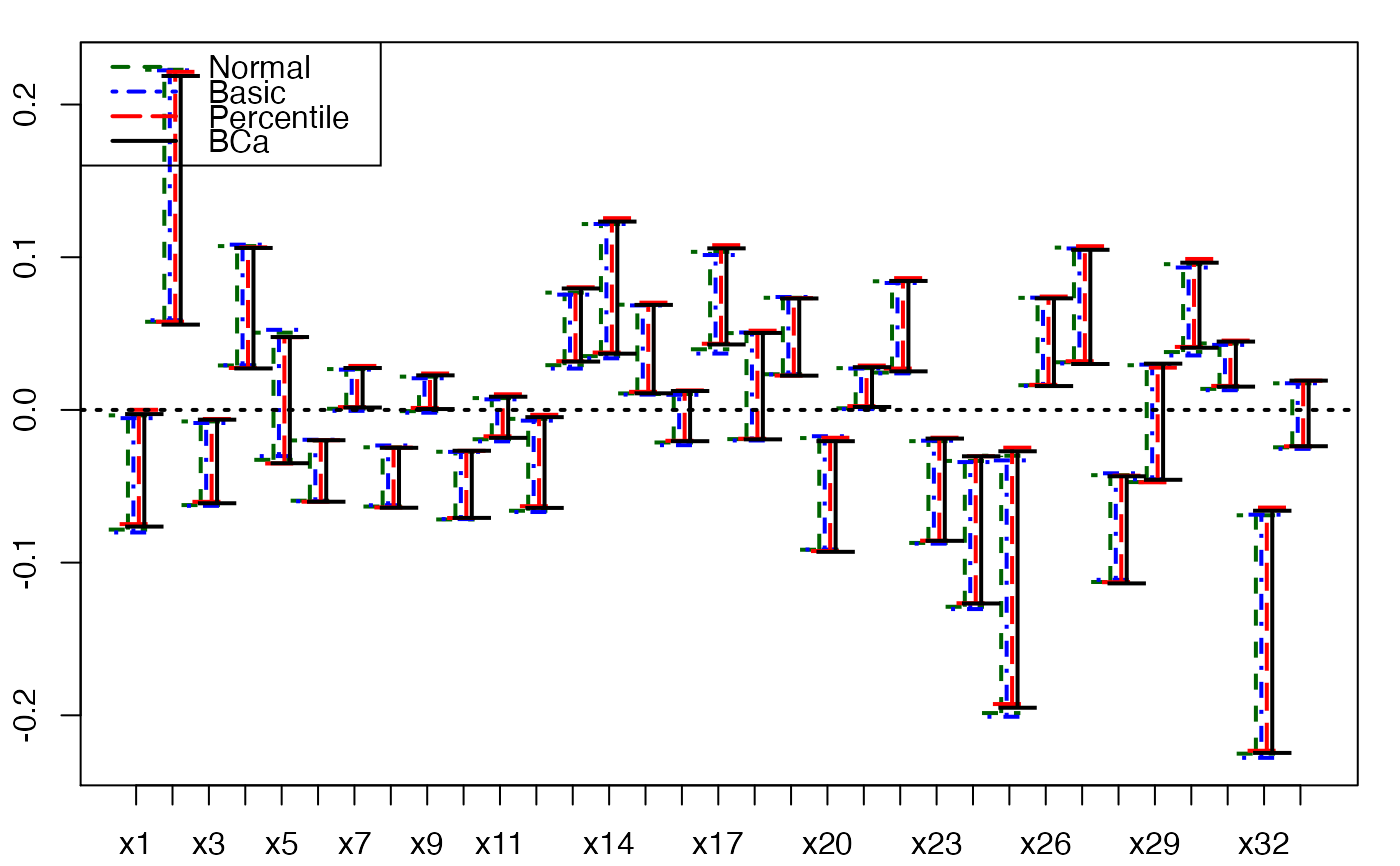 plots.confints.bootpls(temp.ci,c(2,4,6),"bottomleft")
plots.confints.bootpls(temp.ci,c(2,4,6),"bottomleft")
 plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
plots.confints.bootpls(temp.ci,c(2,4,6),articlestyle=FALSE,
main="Bootstrap confidence intervals for some of the bj")
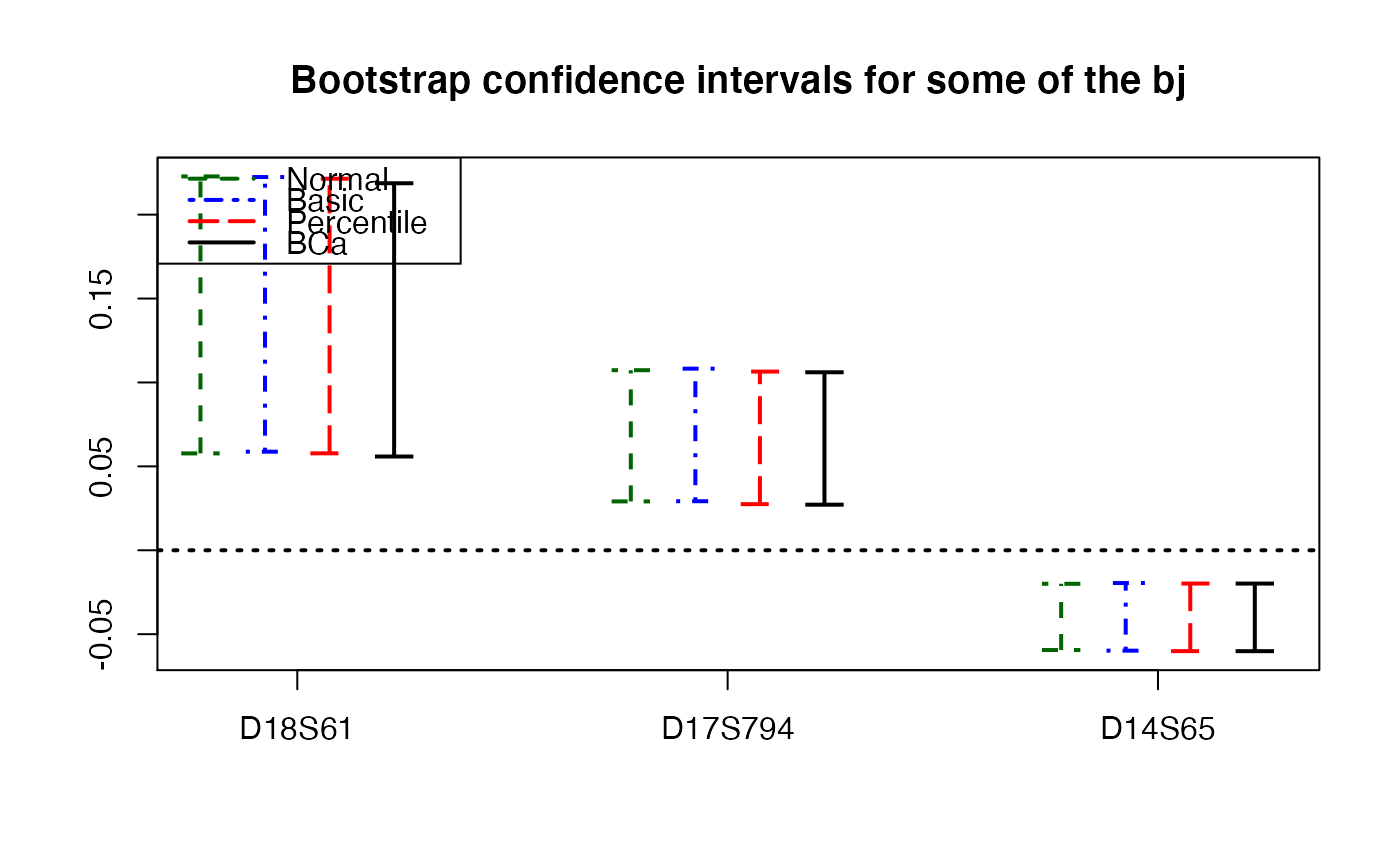 plots.confints.bootpls(temp.ci,prednames=FALSE,ltyIC=c(2,1),colIC=c(1,2))
plots.confints.bootpls(temp.ci,prednames=FALSE,ltyIC=c(2,1),colIC=c(1,2))
 temp.ci <- confints.bootpls(aze_compl.bootYT3,1:33,typeBCa=FALSE)
plots.confints.bootpls(temp.ci,prednames=FALSE)
temp.ci <- confints.bootpls(aze_compl.bootYT3,1:33,typeBCa=FALSE)
plots.confints.bootpls(temp.ci,prednames=FALSE)
 # }
# }
